Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation
- PMID: 18784653
- PMCID: PMC2753212
- DOI: 10.1038/nature07267
Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation
Abstract
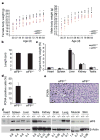
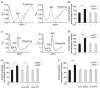
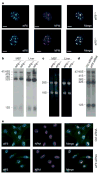
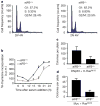
Cell growth and proliferation require coordinated ribosomal biogenesis and translation. Eukaryotic initiation factors (eIFs) control translation at the rate-limiting step of initiation. So far, only two eIFs connect extracellular stimuli to global translation rates: eIF4E acts in the eIF4F complex and regulates binding of capped messenger RNA to 40S subunits, downstream of growth factors, and eIF2 controls loading of the ternary complex on the 40S subunit and is inhibited on stress stimuli. No eIFs have been found to link extracellular stimuli to the activity of the large 60S ribosomal subunit. eIF6 binds 60S ribosomes precluding ribosome joining in vitro. However, studies in yeasts showed that eIF6 is required for ribosome biogenesis rather than translation. Here we show that mammalian eIF6 is required for efficient initiation of translation, in vivo. eIF6 null embryos are lethal at preimplantation. Heterozygous mice have 50% reduction of eIF6 levels in all tissues, and show reduced mass of hepatic and adipose tissues due to a lower number of cells and to impaired G1/S cell cycle progression. eIF6(+/-) cells retain sufficient nucleolar eIF6 and normal ribosome biogenesis. The liver of eIF6(+/-) mice displays an increase of 80S in polysomal profiles, indicating a defect in initiation of translation. Consistently, isolated hepatocytes have impaired insulin-stimulated translation. Heterozygous mouse embryonic fibroblasts recapitulate the organism phenotype and have normal ribosome biogenesis, reduced insulin-stimulated translation, and delayed G1/S phase progression. Furthermore, eIF6(+/-) cells are resistant to oncogene-induced transformation. Thus, eIF6 is the first eIF associated with the large 60S subunit that regulates translation in response to extracellular signals.
Conflict of interest statement
The authors have no competing interests.
Figures
References
-
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. - PubMed
-
- Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. - PubMed
-
- Sonenberg N, Pause A. Signal transduction. Protein synthesis and oncogenesis meet again. Science. 2006;314:428–429. - PubMed
-
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

