Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins
- PMID: 18784751
- PMCID: PMC2567413
- DOI: 10.1038/emboj.2008.184
Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins
Abstract
The LEAFY (LFY) protein is a key regulator of flower development in angiosperms. Its gradually increased expression governs the sharp floral transition, and LFY subsequently controls the patterning of flower meristems by inducing the expression of floral homeotic genes. Despite a wealth of genetic data, how LFY functions at the molecular level is poorly understood. Here, we report crystal structures for the DNA-binding domain of Arabidopsis thaliana LFY bound to two target promoter elements. LFY adopts a novel seven-helix fold that binds DNA as a cooperative dimer, forming base-specific contacts in both the major and minor grooves. Cooperativity is mediated by two basic residues and plausibly accounts for LFY's effectiveness in triggering sharp developmental transitions. Our structure reveals an unexpected similarity between LFY and helix-turn-helix proteins, including homeodomain proteins known to regulate morphogenesis in higher eukaryotes. The appearance of flowering plants has been linked to the molecular evolution of LFY. Our study provides a unique framework to elucidate the molecular mechanisms underlying floral development and the evolutionary history of flowering plants.
Figures

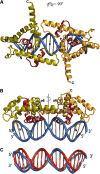



Similar articles
-
A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana.Plant J. 2012 Sep;71(5):736-49. doi: 10.1111/j.1365-313X.2012.05026.x. Epub 2012 Jun 12. Plant J. 2012. PMID: 22507399
-
A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1.Plant J. 2013 May;74(4):678-89. doi: 10.1111/tpj.12156. Epub 2013 Apr 22. Plant J. 2013. PMID: 23445516
-
The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation.Int J Dev Biol. 2012;56(4):207-21. doi: 10.1387/ijdb.113450ns. Int J Dev Biol. 2012. PMID: 22451042 Review.
-
SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis.Curr Biol. 2002 Jan 22;12(2):85-94. doi: 10.1016/s0960-9822(01)00651-0. Curr Biol. 2002. PMID: 11818058
-
[Regulation network and biological roles of LEAFY in Arabidopsis thaliana in floral development].Yi Chuan. 2004 Jan;26(1):137-42. Yi Chuan. 2004. PMID: 15626683 Review. Chinese.
Cited by
-
Enhancing drought tolerance in cauliflower (Brassica oleracea var. botrytis) by targeting LFY transcription factor modulation via the ethylene precursor, ACCA: an innovative computational approach.Front Plant Sci. 2024 Feb 28;15:1255979. doi: 10.3389/fpls.2024.1255979. eCollection 2024. Front Plant Sci. 2024. PMID: 38481405 Free PMC article.
-
Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor.Plant Cell. 2011 Apr;23(4):1293-306. doi: 10.1105/tpc.111.083329. Epub 2011 Apr 22. Plant Cell. 2011. PMID: 21515819 Free PMC article.
-
Transcription Factor Interplay between LEAFY and APETALA1/CAULIFLOWER during Floral Initiation.Plant Physiol. 2017 Jun;174(2):1097-1109. doi: 10.1104/pp.17.00098. Epub 2017 Apr 6. Plant Physiol. 2017. PMID: 28385730 Free PMC article.
-
The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele.Plants (Basel). 2019 Dec 13;8(12):599. doi: 10.3390/plants8120599. Plants (Basel). 2019. PMID: 31847079 Free PMC article.
-
Evolution of the Plant Reproduction Master Regulators LFY and the MADS Transcription Factors: The Role of Protein Structure in the Evolutionary Development of the Flower.Front Plant Sci. 2016 Jan 6;6:1193. doi: 10.3389/fpls.2015.01193. eCollection 2015. Front Plant Sci. 2016. PMID: 26779227 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Albert VA, Oppenheimer DG, Lindqvist C (2002) Pleiotropy, redundancy and the evolution of flowers. Trends Plant Sci 7: 297–301 - PubMed
-
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 - PubMed
-
- Blazquez MA, Ferrandiz C, Madueno F, Parcy F (2006) How floral meristems are built. Plant Mol Biol 60: 855–870 - PubMed
-
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J (2003) Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

