Functional copy-number alterations in cancer
- PMID: 18784837
- PMCID: PMC2527508
- DOI: 10.1371/journal.pone.0003179
Functional copy-number alterations in cancer
Abstract
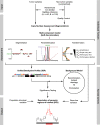
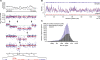
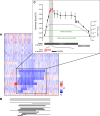
Understanding the molecular basis of cancer requires characterization of its genetic defects. DNA microarray technologies can provide detailed raw data about chromosomal aberrations in tumor samples. Computational analysis is needed (1) to deduce from raw array data actual amplification or deletion events for chromosomal fragments and (2) to distinguish causal chromosomal alterations from functionally neutral ones. We present a comprehensive computational approach, RAE, designed to robustly map chromosomal alterations in tumor samples and assess their functional importance in cancer. To demonstrate the methodology, we experimentally profile copy number changes in a clinically aggressive subtype of soft-tissue sarcoma, pleomorphic liposarcoma, and computationally derive a portrait of candidate oncogenic alterations and their target genes. Many affected genes are known to be involved in sarcomagenesis; others are novel, including mediators of adipocyte differentiation, and may include valuable therapeutic targets. Taken together, we present a statistically robust methodology applicable to high-resolution genomic data to assess the extent and function of copy-number alterations in cancer.
Conflict of interest statement
Figures
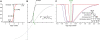

Similar articles
-
"Lineage addiction" in human cancer: lessons from integrated genomics.Cold Spring Harb Symp Quant Biol. 2005;70:25-34. doi: 10.1101/sqb.2005.70.016. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869735
-
Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number.Cancer Res. 2006 Jun 15;66(12):6050-62. doi: 10.1158/0008-5472.CAN-05-4618. Cancer Res. 2006. PMID: 16778177
-
TAFFYS: An Integrated Tool for Comprehensive Analysis of Genomic Aberrations in Tumor Samples.PLoS One. 2015 Jun 25;10(6):e0129835. doi: 10.1371/journal.pone.0129835. eCollection 2015. PLoS One. 2015. PMID: 26111017 Free PMC article.
-
Existing and emerging technologies for tumor genomic profiling.J Clin Oncol. 2013 May 20;31(15):1815-24. doi: 10.1200/JCO.2012.46.5948. Epub 2013 Apr 15. J Clin Oncol. 2013. PMID: 23589546 Free PMC article. Review.
-
Understanding genomic alterations in cancer genomes using an integrative network approach.Cancer Lett. 2013 Nov 1;340(2):261-9. doi: 10.1016/j.canlet.2012.11.050. Epub 2012 Dec 22. Cancer Lett. 2013. PMID: 23266571 Review.
Cited by
-
Identification of tumor suppressors and oncogenes from genomic and epigenetic features in ovarian cancer.PLoS One. 2011;6(12):e28503. doi: 10.1371/journal.pone.0028503. Epub 2011 Dec 8. PLoS One. 2011. PMID: 22174824 Free PMC article.
-
Proteomic research in sarcomas - current status and future opportunities.Semin Cancer Biol. 2020 Apr;61:56-70. doi: 10.1016/j.semcancer.2019.11.003. Epub 2019 Nov 10. Semin Cancer Biol. 2020. PMID: 31722230 Free PMC article. Review.
-
Integrative genomic profiling of human prostate cancer.Cancer Cell. 2010 Jul 13;18(1):11-22. doi: 10.1016/j.ccr.2010.05.026. Epub 2010 Jun 24. Cancer Cell. 2010. PMID: 20579941 Free PMC article.
-
Soft tissue sarcomas with complex genomic profiles.Virchows Arch. 2010 Feb;456(2):201-17. doi: 10.1007/s00428-009-0853-4. Virchows Arch. 2010. PMID: 20217954 Review.
-
Establishment of highly tumorigenic human colorectal cancer cell line (CR4) with properties of putative cancer stem cells.PLoS One. 2014 Jun 12;9(6):e99091. doi: 10.1371/journal.pone.0099091. eCollection 2014. PLoS One. 2014. PMID: 24921652 Free PMC article.
References
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
-
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. - PubMed
-
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

