Functional characteristics of a highly specific integrase encoded by an LTR-retrotransposon
- PMID: 18784842
- PMCID: PMC2527525
- DOI: 10.1371/journal.pone.0003185
Functional characteristics of a highly specific integrase encoded by an LTR-retrotransposon
Abstract
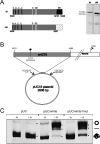
Background: The retroviral Integrase protein catalyzes the insertion of linear viral DNA into host cell DNA. Although different retroviruses have been shown to target distinctive chromosomal regions, few of them display a site-specific integration. ZAM, a retroelement from Drosophila melanogaster very similar in structure and replication cycle to mammalian retroviruses is highly site-specific. Indeed, ZAM copies target the genomic 5'-CGCGCg-3' consensus-sequences. To enlighten the determinants of this high integration specificity, we investigated the functional properties of its integrase protein denoted ZAM-IN.
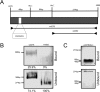
Principal findings: Here we show that ZAM-IN displays the property to nick DNA molecules in vitro. This endonuclease activity targets specific sequences that are present in a 388 bp fragment taken from the white locus and known to be a genomic ZAM integration site in vivo. Furthermore, ZAM-IN displays the unusual property to directly bind specific genomic DNA sequences. Two specific and independent sites are recognized within the 388 bp fragment of the white locus: the CGCGCg sequence and a closely apposed site different in sequence.
Conclusion: This study strongly argues that the intrinsic properties of ZAM-IN, ie its binding properties and its endonuclease activity, play an important part in ZAM integration specificity. Its ability to select two binding sites and to nick the DNA molecule reminds the strategy used by some site-specific recombination enzymes and forms the basis for site-specific integration strategies potentially useful in a broad range of genetic engineering applications.
Conflict of interest statement
Figures


Similar articles
-
The integration machinery of ZAM, a retroelement from Drosophila melanogaster, acts as a sequence-specific endonuclease.J Virol. 1999 Aug;73(8):7061-4. doi: 10.1128/JVI.73.8.7061-7064.1999. J Virol. 1999. PMID: 10400810 Free PMC article.
-
Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster.Viruses. 2017 Apr 16;9(4):81. doi: 10.3390/v9040081. Viruses. 2017. PMID: 28420154 Free PMC article. Review.
-
Integration specificity of LTR-retrotransposons and retroviruses in the Drosophila melanogaster genome.Virus Genes. 2011 Apr;42(2):297-306. doi: 10.1007/s11262-010-0566-4. Epub 2011 Jan 8. Virus Genes. 2011. PMID: 21369828
-
[Precise excision of long terminal repeats of the gypsy (mdg4) retrotransposon of Drosophila melanogaster detected in Escherichia coli cells is explained by its integrase function].Genetika. 2006 Dec;42(12):1656-63. Genetika. 2006. PMID: 17326385 Russian.
-
Impact of multiple insertions of two retroelements, ZAM and Idefix at an euchromatic locus.Genetica. 2000;109(1-2):53-9. doi: 10.1023/a:1026534207401. Genetica. 2000. PMID: 11293795 Review.
Cited by
-
Integration site selection by retroviruses and transposable elements in eukaryotes.Nat Rev Genet. 2017 May;18(5):292-308. doi: 10.1038/nrg.2017.7. Epub 2017 Mar 13. Nat Rev Genet. 2017. PMID: 28286338 Review.
References
-
- Coffin JM, Hughes SH, Varmus HE, Vogt PK, Vogt VM, et al. Retrovirus Edited by J. M. Coffin, S. H. Hugues and H. E. Varmus. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997.
-
- Lewinski MK, Bushman FD. Retroviral DNA integration–mechanism and consequences. Adv Genet. 2005;55:147–181. - PubMed
-
- Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

