Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization
- PMID: 18785738
- PMCID: PMC3195406
- DOI: 10.1021/ja802215c
Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization
Abstract
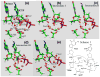
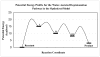
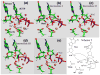
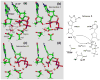
The nucleotidyl-transfer reaction coupled with the conformational transitions in DNA polymerases is critical for maintaining the fidelity and efficiency of DNA synthesis. We examine here the possible reaction pathways of a Y-family DNA polymerase, Sulfolobus solfataricus DNA polymerase IV (Dpo4), for the correct insertion of dCTP opposite 8-oxoguanine using the quantum mechanics/molecular mechanics (QM/MM) approach, both from a chemistry-competent state and a crystal closed state. The latter examination is important for understanding pre-chemistry barriers to interpret the entire enzyme mechanism, since the crystal closed state is not an ideal state for initiating the chemical reaction. The most favorable reaction path involves initial deprotonation of O3'H via two bridging water molecules to O1A, overcoming an overall potential energy barrier of approximately 20.0 kcal/mol. The proton on O1A-P(alpha) then migrates to the gamma-phosphate oxygen of the incoming nucleotide as O3' attacks P(alpha), and the P(alpha)-O3A bond breaks. The other possible pathway in which the O3'H proton is transferred directly to O1A on P(alpha) has an overall energy barrier of 25.0 kcal/mol. In both reaction paths, the rate-limiting step is the initial deprotonation, and the trigonal-bipyramidal configuration for P(alpha) occurs during the concerted bond formation (O3'-P(alpha)) and breaking (P(alpha)-O3A), indicating the associative nature of the chemical reaction. In contrast, the Dpo4/DNA complex with an imperfect active-site geometry corresponding to the crystal state must overcome a much higher activation energy barrier (29.0 kcal/mol) to achieve a tightly organized site due to hindered O3'H deprotonation stemming from larger distances and distorted conformation of the proton acceptors. This significant difference demonstrates that the pre-chemistry reorganization in Dpo4 costs approximately 4.0 to 9.0 kcal/mol depending on the primer terminus environment. Compared to the higher fidelity DNA polymerase beta from the X-family, Dpo4 has a higher chemical reaction barrier (20.0 vs 15.0 kcal/mol) due to the more solvent-exposed active site.
Figures




Similar articles
-
Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase κ elucidates error-free bypass of a bulky DNA lesion.Nucleic Acids Res. 2012 Oct;40(18):9193-205. doi: 10.1093/nar/gks653. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772988 Free PMC article.
-
A quantum mechanical investigation of possible mechanisms for the nucleotidyl transfer reaction catalyzed by DNA polymerase beta.J Phys Chem B. 2007 Sep 27;111(38):11244-52. doi: 10.1021/jp071838c. Epub 2007 Sep 1. J Phys Chem B. 2007. PMID: 17764165
-
DNA polymerase beta catalysis: are different mechanisms possible?J Am Chem Soc. 2007 Sep 12;129(36):11100-10. doi: 10.1021/ja071533b. Epub 2007 Aug 16. J Am Chem Soc. 2007. PMID: 17696533
-
Regulation of DNA repair fidelity by molecular checkpoints: "gates" in DNA polymerase beta's substrate selection.Biochemistry. 2006 Dec 26;45(51):15142-56. doi: 10.1021/bi061353z. Epub 2006 Dec 1. Biochemistry. 2006. PMID: 17176036 Free PMC article. Review.
-
Applications of quantum mechanical/molecular mechanical methods to the chemical insertion step of DNA and RNA polymerization.Adv Protein Chem Struct Biol. 2014;97:83-113. doi: 10.1016/bs.apcsb.2014.10.001. Epub 2014 Nov 7. Adv Protein Chem Struct Biol. 2014. PMID: 25458356 Free PMC article. Review.
Cited by
-
Computational delineation of the catalytic step of a high-fidelity DNA polymerase.Protein Sci. 2010 Apr;19(4):815-25. doi: 10.1002/pro.361. Protein Sci. 2010. PMID: 20162624 Free PMC article.
-
Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase κ elucidates error-free bypass of a bulky DNA lesion.Nucleic Acids Res. 2012 Oct;40(18):9193-205. doi: 10.1093/nar/gks653. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772988 Free PMC article.
-
Ultrafast water dynamics at the interface of the polymerase-DNA binding complex.Biochemistry. 2014 Aug 26;53(33):5405-13. doi: 10.1021/bi500810a. Epub 2014 Aug 15. Biochemistry. 2014. PMID: 25105470 Free PMC article.
-
Computational Simulations of DNA Polymerases: Detailed Insights on Structure/Function/Mechanism from Native Proteins to Cancer Variants.Chem Res Toxicol. 2017 Nov 20;30(11):1922-1935. doi: 10.1021/acs.chemrestox.7b00161. Epub 2017 Sep 15. Chem Res Toxicol. 2017. PMID: 28877429 Free PMC article.
-
Addressing open questions about phosphate hydrolysis pathways by careful free energy mapping.J Phys Chem B. 2013 Jan 10;117(1):153-63. doi: 10.1021/jp309778n. Epub 2012 Dec 28. J Phys Chem B. 2013. PMID: 23198768 Free PMC article.
References
-
- Friedberg EC. DNA damage and repair. Nature. 2003;421(6921):436–440. - PubMed
-
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. - PubMed
-
- Ohmori H, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8(1):7–8. - PubMed
-
- Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion: Structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

