Localization of general and regulatory proteolysis in Bacillus subtilis cells
- PMID: 18786145
- PMCID: PMC2628427
- DOI: 10.1111/j.1365-2958.2008.06438.x
Localization of general and regulatory proteolysis in Bacillus subtilis cells
Abstract
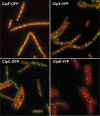
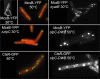
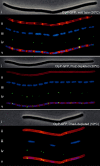
Protein degradation mediated by ATP-dependent proteases, such as Hsp100/Clp and related AAA+ proteins, plays an important role in cellular protein homeostasis, protein quality control and the regulation of, e.g. heat shock adaptation and other cellular differentiation processes. ClpCP with its adaptor proteins and other related proteases, such as ClpXP or ClpEP of Bacillus subtilis, are involved in general and regulatory proteolysis. To determine if proteolysis occurs at specific locations in B. subtilis cells, we analysed the subcellular distribution of the Clp system together with adaptor and general and regulatory substrate proteins, under different environmental conditions. We can demonstrate that the ATPase and the proteolytic subunit of the Clp proteases, as well as the adaptor or substrate proteins, form visible foci, representing active protease clusters localized to the polar and to the mid-cell region. These clusters could represent a compartmentalized place for protein degradation positioned at the pole close to where most of the cellular protein biosynthesis and also protein quality control are taking place, thereby spatially separating protein synthesis and degradation.
Figures




Similar articles
-
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.Nat Rev Microbiol. 2009 Aug;7(8):589-99. doi: 10.1038/nrmicro2185. Nat Rev Microbiol. 2009. PMID: 19609260 Review.
-
Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis.Res Microbiol. 2009 Nov;160(9):637-44. doi: 10.1016/j.resmic.2009.08.020. Epub 2009 Sep 23. Res Microbiol. 2009. PMID: 19781636 Review.
-
Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis.J Bacteriol. 2008 Oct;190(20):6749-57. doi: 10.1128/JB.00589-08. Epub 2008 Aug 8. J Bacteriol. 2008. PMID: 18689476 Free PMC article.
-
The tyrosine kinase McsB is a regulated adaptor protein for ClpCP.EMBO J. 2007 Apr 18;26(8):2061-70. doi: 10.1038/sj.emboj.7601655. Epub 2007 Mar 22. EMBO J. 2007. PMID: 17380125 Free PMC article.
-
Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis.J Bacteriol. 2008 Oct;190(20):6758-68. doi: 10.1128/JB.00590-08. Epub 2008 Aug 8. J Bacteriol. 2008. PMID: 18689473 Free PMC article.
Cited by
-
Activity control of the ClpC adaptor McsB in Bacillus subtilis.J Bacteriol. 2011 Aug;193(15):3887-93. doi: 10.1128/JB.00079-11. Epub 2011 May 27. J Bacteriol. 2011. PMID: 21622759 Free PMC article.
-
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.Nat Rev Microbiol. 2009 Aug;7(8):589-99. doi: 10.1038/nrmicro2185. Nat Rev Microbiol. 2009. PMID: 19609260 Review.
-
Integrating protein homeostasis strategies in prokaryotes.Cold Spring Harb Perspect Biol. 2011 Apr 1;3(4):a004366. doi: 10.1101/cshperspect.a004366. Cold Spring Harb Perspect Biol. 2011. PMID: 21441580 Free PMC article. Review.
-
The trpE gene negatively regulates differentiation of heterocysts at the level of induction in Anabaena sp. strain PCC 7120.J Bacteriol. 2015 Jan;197(2):362-70. doi: 10.1128/JB.02145-14. Epub 2014 Nov 10. J Bacteriol. 2015. PMID: 25384479 Free PMC article.
-
Protein aggregation in bacteria.FEMS Microbiol Rev. 2020 Jan 1;44(1):54-72. doi: 10.1093/femsre/fuz026. FEMS Microbiol Rev. 2020. PMID: 31633151 Free PMC article. Review.
References
-
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

