The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets
- PMID: 18786525
- PMCID: PMC3137263
- DOI: 10.1016/j.ydbio.2008.08.011
The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets
Abstract
The highly conserved Wingless/Wnt signaling pathway controls many developmental processes by regulating the expression of target genes, most often through members of the TCF family of DNA-binding proteins. In the absence of signaling, many of these targets are silenced, by mechanisms involving TCFs that are not fully understood. Here we report that the chromatin remodeling proteins ISWI and ACF1 are required for basal repression of WG target genes in Drosophila. This regulation is not due to global repression by ISWI and ACF1 and is distinct from their previously reported role in chromatin assembly. While ISWI is localized to the same regions of Wingless target gene chromatin as TCF, we find that ACF1 binds much more broadly to target loci. This broad distribution of ACF1 is dependent on ISWI. ISWI and ACF1 are required for TCF binding to chromatin, while a TCF-independent role of ISWI-ACF1 in repression of Wingless targets is also observed. Finally, we show that Wingless signaling reduces ACF1 binding to WG targets, and ISWI and ACF1 regulate repression by antagonizing histone H4 acetylation. Our results argue that WG signaling activates target gene expression partly by overcoming the chromatin barrier maintained by ISWI and ACF1.
Figures

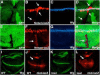



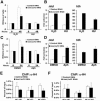
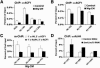
Similar articles
-
The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway.EMBO Rep. 2009 Oct;10(10):1140-6. doi: 10.1038/embor.2009.157. Epub 2009 Aug 28. EMBO Rep. 2009. PMID: 19713963 Free PMC article.
-
ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly.Genes Dev. 1999 Jun 15;13(12):1529-39. doi: 10.1101/gad.13.12.1529. Genes Dev. 1999. PMID: 10385622 Free PMC article.
-
Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo.Genes Dev. 2004 Jan 15;18(2):170-83. doi: 10.1101/gad.1139604. Genes Dev. 2004. PMID: 14752009 Free PMC article.
-
Modulating and measuring Wingless signalling.Methods. 2014 Jun 15;68(1):194-8. doi: 10.1016/j.ymeth.2014.03.015. Epub 2014 Mar 25. Methods. 2014. PMID: 24675402 Review.
-
Imitation switch complexes.Ernst Schering Res Found Workshop. 2006;(57):61-87. doi: 10.1007/3-540-37633-x_4. Ernst Schering Res Found Workshop. 2006. PMID: 16568949 Review.
Cited by
-
Divergent human remodeling complexes remove nucleosomes from strong positioning sequences.Nucleic Acids Res. 2010 Jan;38(2):400-13. doi: 10.1093/nar/gkp1030. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906705 Free PMC article.
-
ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior.Nat Med. 2015 Oct;21(10):1146-53. doi: 10.1038/nm.3939. Epub 2015 Sep 21. Nat Med. 2015. PMID: 26390241 Free PMC article.
-
The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences.Nucleic Acids Res. 2015 Feb 18;43(3):1444-55. doi: 10.1093/nar/gku1371. Epub 2015 Jan 10. Nucleic Acids Res. 2015. PMID: 25578963 Free PMC article.
-
Beta-catenin hits chromatin: regulation of Wnt target gene activation.Nat Rev Mol Cell Biol. 2009 Apr;10(4):276-86. doi: 10.1038/nrm2654. Nat Rev Mol Cell Biol. 2009. PMID: 19305417 Review.
-
NLP is a novel transcription regulator involved in VSG expression site control in Trypanosoma brucei.Nucleic Acids Res. 2011 Mar;39(6):2018-31. doi: 10.1093/nar/gkq950. Epub 2010 Nov 13. Nucleic Acids Res. 2011. PMID: 21076155 Free PMC article.
References
-
- Azpiazu N, Morata G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development. 2000;127:2685–2693. - PubMed
-
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

