Transcriptome analysis identifies genes with enriched expression in the mouse central extended amygdala
- PMID: 18786617
- PMCID: PMC2629946
- DOI: 10.1016/j.neuroscience.2008.07.070
Transcriptome analysis identifies genes with enriched expression in the mouse central extended amygdala
Abstract
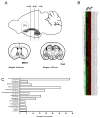
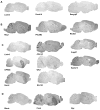
The central extended amygdala (EAc) is an ensemble of highly interconnected limbic structures of the anterior brain, and forms a cellular continuum including the bed nucleus of the stria terminalis (BNST), the central nucleus of the amygdala (CeA) and the nucleus accumbens shell (AcbSh). This neural network is a key site for interactions between brain reward and stress systems, and has been implicated in several aspects of drug abuse. In order to increase our understanding of EAc function at the molecular level, we undertook a genome-wide screen (Affymetrix) to identify genes whose expression is enriched in the mouse EAc. We focused on the less-well known BNST-CeA areas of the EAc, and identified 121 genes that exhibit more than twofold higher expression level in the EAc compared with whole brain. Among these, 43 genes have never been described to be expressed in the EAc. We mapped these genes throughout the brain, using non-radioactive in situ hybridization, and identified eight genes with a unique and distinct rostro-caudal expression pattern along AcbSh, BNST and CeA. Q-PCR analysis performed in brain and peripheral organ tissues indicated that, with the exception of one (Spata13), all these genes are predominantly expressed in brain. These genes encode signaling proteins (Adora2, GPR88, Arpp21 and Rem2), a transcription factor (Limh6) or proteins of unknown function (Rik130, Spata13 and Wfs1). The identification of genes with enriched expression expands our knowledge of EAc at a molecular level, and provides useful information to toward genetic manipulations within the EAc.
Figures




References
-
- Abou-Sleymane G, Chalmel F, Helmlinger D, Lardenois A, Thibault C, Weber C, Merienne K, Mandel JL, Poch O, Devys D, Trottier Y. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum Mol Genet. 2006;15:691–703. - PubMed
-
- Alheid GF, Heimer L. New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata. Neuroscience. 1988;27:1–39. - PubMed
-
- Befort K, Filliol D, Darcq E, Ghate A, Matifas A, Lardenois A, Muller J, Thibault C, Dembele D, Poch O, Kieffer BL. Gene expression is altered in the lateral hypothalamus upon activation of the mu opioid receptor. Ann N Y Acad Sci. 2008a;1129:175–184. - PubMed
-
- Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, Lardenois A, Thibault C, Dembele D, Le Merrer J, Becker JAJ, Poch O, Kieffer BL. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. J neurosci. 2008b;27:2973–2984. - PubMed
-
- Bonaventure P, Guo H, Tian B, Liu X, Bittner A, Roland B, Salunga R, Ma XJ, Kamme F, Meurers B, Bakker M, Jurzak M, Leysen JE, Erlander MG. Nuclei and subnuclei gene expression profiling in mammalian brain. Brain Res. 2002;943:38–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

