Folding and unfolding single RNA molecules under tension
- PMID: 18786653
- PMCID: PMC2855187
- DOI: 10.1016/j.cbpa.2008.08.011
Folding and unfolding single RNA molecules under tension
Abstract
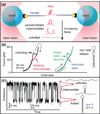
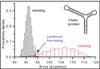
Single-molecule force spectroscopy constitutes a powerful method for probing RNA folding: It allows the kinetic, energetic, and structural properties of intermediate and transition states to be determined quantitatively, yielding new insights into folding pathways and energy landscapes. Recent advances in experimental and theoretical methods, including fluctuation theorems, kinetic theories, novel force clamps, and ultrastable instruments, have opened new avenues for study. These tools have been used to probe folding in simple model systems, for example, RNA and DNA hairpins. Knowledge gained from such systems is helping to build our understanding of more complex RNA structures composed of multiple elements, as well as how nucleic acids interact with proteins involved in key cellular activities, such as transcription and translation.
Figures


Similar articles
-
Unfolding and melting of DNA (RNA) hairpins: the concept of structure-specific 2D dynamic landscapes.Phys Chem Chem Phys. 2008 Aug 7;10(29):4227-39. doi: 10.1039/b804675c. Epub 2008 Jun 3. Phys Chem Chem Phys. 2008. PMID: 18633543 Free PMC article.
-
Mechanical unfolding of RNA: from hairpins to structures with internal multiloops.Biophys J. 2007 Feb 1;92(3):731-43. doi: 10.1529/biophysj.106.093062. Epub 2006 Oct 6. Biophys J. 2007. PMID: 17028142 Free PMC article.
-
Mechanically probing the folding pathway of single RNA molecules.Biophys J. 2003 May;84(5):2831-40. doi: 10.1016/S0006-3495(03)70012-5. Biophys J. 2003. PMID: 12719217 Free PMC article.
-
Exploring RNA folding one molecule at a time.Curr Opin Chem Biol. 2008 Dec;12(6):647-54. doi: 10.1016/j.cbpa.2008.09.010. Epub 2008 Oct 7. Curr Opin Chem Biol. 2008. PMID: 18845269 Review.
-
Structures of helical junctions in nucleic acids.Q Rev Biophys. 2000 May;33(2):109-59. doi: 10.1017/s0033583500003590. Q Rev Biophys. 2000. PMID: 11131562 Review.
Cited by
-
Moving into the cell: single-molecule studies of molecular motors in complex environments.Nat Rev Mol Cell Biol. 2011 Mar;12(3):163-76. doi: 10.1038/nrm3062. Epub 2011 Feb 16. Nat Rev Mol Cell Biol. 2011. PMID: 21326200 Review.
-
Reconstructing folding energy landscapes from splitting probability analysis of single-molecule trajectories.Proc Natl Acad Sci U S A. 2015 Jun 9;112(23):7183-8. doi: 10.1073/pnas.1419490112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26039984 Free PMC article.
-
Determining intrachain diffusion coefficients for biopolymer dynamics from single-molecule force spectroscopy measurements.Biophys J. 2014 Oct 7;107(7):1647-53. doi: 10.1016/j.bpj.2014.08.007. Biophys J. 2014. PMID: 25296317 Free PMC article.
-
High Spatiotemporal-Resolution Magnetic Tweezers: Calibration and Applications for DNA Dynamics.Biophys J. 2015 Nov 17;109(10):2113-25. doi: 10.1016/j.bpj.2015.10.018. Biophys J. 2015. PMID: 26588570 Free PMC article.
-
Compact intermediates in RNA folding.Annu Rev Biophys. 2010;39:61-77. doi: 10.1146/annurev.biophys.093008.131334. Annu Rev Biophys. 2010. PMID: 20192764 Free PMC article. Review.
References
-
- Brion P, Westhof E. Hierarchy and dynamics. Annu Rev Biophys Biomol Struct. 1997;26:113–137. - PubMed
-
- Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. - PubMed
-
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical Processes in Biochemistry. Annu Rev Biochem. 2004;73:705–748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

