Hypermutation at A/T sites during G.U mismatch repair in vitro by human B-cell lysates
- PMID: 18786917
- PMCID: PMC2581569
- DOI: 10.1074/jbc.M805524200
Hypermutation at A/T sites during G.U mismatch repair in vitro by human B-cell lysates
Abstract
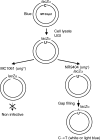
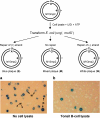
Somatic hypermutation in the variable regions of immunoglobulin genes is required to produce high affinity antibody molecules. Somatic hypermutation results by processing G.U mismatches generated when activation-induced cytidine deaminase (AID) deaminates C to U. Mutations at C/G sites are targeted mainly at deamination sites, whereas mutations at A/T sites entail error-prone DNA gap repair. We used B-cell lysates to analyze salient features of somatic hypermutation with in vitro mutational assays. Tonsil and hypermutating Ramos B-cells convert C-->U in accord with AID motif specificities, whereas HeLa cells do not. Using tonsil cell lysates to repair a G.U mismatch, A/T and G/C targeted mutations occur about equally, whereas Ramos cell lysates make fewer mutations at A/T sites (approximately 24%) compared with G/C sites (approximately 76%). In contrast, mutations in HeLa cell lysates occur almost exclusively at G/C sites (> 95%). By recapitulating two basic features of B-cell-specific somatic hypermutation, G/C mutations targeted to AID hot spot motifs and elevated A/T mutations dependent on error-prone processing of G.U mispairs, these cell free assays provide a practical method to reconstitute error-prone mismatch repair using purified B-cell proteins.
Figures

Similar articles
-
Analysis of Ig gene hypermutation in Ung(-/-)Polh(-/-) mice suggests that UNG and A:T mutagenesis pathway target different U:G lesions.Mol Immunol. 2013 Mar;53(3):214-7. doi: 10.1016/j.molimm.2012.08.009. Epub 2012 Sep 4. Mol Immunol. 2013. PMID: 22960197
-
Integrity of immunoglobulin variable regions is supported by GANP during AID-induced somatic hypermutation in germinal center B cells.Int Immunol. 2017 May 1;29(5):211-220. doi: 10.1093/intimm/dxx032. Int Immunol. 2017. PMID: 28541550 Free PMC article.
-
Repertoire Sequencing of B Cells Elucidates the Role of UNG and Mismatch Repair Proteins in Somatic Hypermutation in Humans.Front Immunol. 2019 Aug 27;10:1913. doi: 10.3389/fimmu.2019.01913. eCollection 2019. Front Immunol. 2019. PMID: 31507588 Free PMC article.
-
Towards an understanding of somatic hypermutation.Curr Opin Immunol. 2001 Apr;13(2):208-18. doi: 10.1016/s0952-7915(00)00206-5. Curr Opin Immunol. 2001. PMID: 11228415 Review.
-
The biochemistry of somatic hypermutation.Annu Rev Immunol. 2008;26:481-511. doi: 10.1146/annurev.immunol.26.021607.090236. Annu Rev Immunol. 2008. PMID: 18304001 Review.
Cited by
-
Decreased mutation frequencies among immunoglobulin G variable region genes during viremic HIV-1 infection.PLoS One. 2014 Jan 7;9(1):e81913. doi: 10.1371/journal.pone.0081913. eCollection 2014. PLoS One. 2014. PMID: 24409278 Free PMC article.
-
Biochemical basis of immunological and retroviral responses to DNA-targeted cytosine deamination by activation-induced cytidine deaminase and APOBEC3G.J Biol Chem. 2009 Oct 9;284(41):27761-27765. doi: 10.1074/jbc.R109.052449. Epub 2009 Aug 13. J Biol Chem. 2009. PMID: 19684020 Free PMC article. Review.
-
Immunoglobulin somatic hypermutation by APOBEC3/Rfv3 during retroviral infection.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7759-64. doi: 10.1073/pnas.1403361111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821801 Free PMC article.
-
Histone H2A and H2B are monoubiquitinated at AID-targeted loci.PLoS One. 2010 Jul 16;5(7):e11641. doi: 10.1371/journal.pone.0011641. PLoS One. 2010. PMID: 20661291 Free PMC article.
-
Simultaneous in vitro characterisation of DNA deaminase function and associated DNA repair pathways.PLoS One. 2013 Dec 9;8(12):e82097. doi: 10.1371/journal.pone.0082097. eCollection 2013. PLoS One. 2013. PMID: 24349193 Free PMC article.
References
-
- Rajewsky, K., Forster, I., and Cumano, A. (1987) Science 238 1088-1094 - PubMed
-
- Chaudhuri, J., and Alt, F. W. (2004) Nat. Rev. Immunol. 4 541-552 - PubMed
-
- Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y., and Honjo, T. (2000) Cell 102 553-563 - PubMed
-
- Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., Tezcan, I., Ersoy, F., Kayserili, H., Ugazio, A. G., Brousse, N., Muramatsu, M., Notarangelo, L. D., Kinoshita, K., Honjo, T., Fischer, A., and Durandy, A. (2000) Cell 102 565-575 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

