Multiple virus lineages sharing recent common ancestry were associated with a Large Rift Valley fever outbreak among livestock in Kenya during 2006-2007
- PMID: 18786992
- PMCID: PMC2573244
- DOI: 10.1128/JVI.01519-08
Multiple virus lineages sharing recent common ancestry were associated with a Large Rift Valley fever outbreak among livestock in Kenya during 2006-2007
Abstract
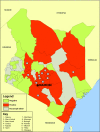
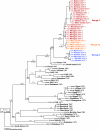
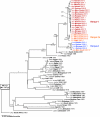
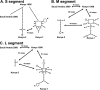
Rift Valley fever (RVF) virus historically has caused widespread and extensive outbreaks of severe human and livestock disease throughout Africa, Madagascar, and the Arabian Peninsula. Following unusually heavy rainfall during the late autumn of 2006, reports of human and animal illness consistent with RVF virus infection emerged across semiarid regions of the Garissa District of northeastern Kenya and southern Somalia. Following initial RVF virus laboratory confirmation, a high-throughput RVF diagnostic facility was established at the Kenyan Central Veterinary Laboratories in Kabete, Kenya, to support the real-time identification of infected livestock and to facilitate outbreak response and control activities. A total of 3,250 specimens from a variety of animal species, including domesticated livestock (cattle, sheep, goats, and camels) and wildlife collected from a total of 55 of 71 Kenyan administrative districts, were tested by molecular and serologic assays. Evidence of RVF infection was found in 9.2% of animals tested and across 23 districts of Kenya, reflecting the large number of affected livestock and the geographic extent of the outbreak. The complete S, M, and/or L genome segment sequence was obtained from a total of 31 RVF virus specimens spanning the entire known outbreak period (December-May) and geographic areas affected by RVF virus activity. Extensive genomic analyses demonstrated the concurrent circulation of multiple virus lineages, gene segment reassortment, and the common ancestry of the 2006/2007 outbreak viruses with those from the 1997-1998 east African RVF outbreak. Evidence of recent increases in genomic diversity and effective population size 2 to 4 years prior to the 2006-2007 outbreak also was found, indicating ongoing RVF virus activity and evolution during the interepizootic/epidemic period. These findings have implications for further studies of basic RVF virus ecology and the design of future surveillance/diagnostic activities, and they highlight the critical need for safe and effective vaccines and antiviral compounds to combat this significant veterinary and public health threat.
Figures


References
-
- Anonymous. 2007. Outbreaks of Rift Valley fever in Kenya, Somalia and United Republic of Tanzania, December 2006-April 2007. Wkly. Epidemiol. Rec. 82169-178. - PubMed
-
- Beaty, B. J., E. J. Rozhon, P. Gensemer, and D. H. Bishop. 1981. Formation of reassortant bunyaviruses in dually infected mosquitoes. Virology 111662-665. - PubMed
-
- Bird, B. H., C. G. Albarino, and S. T. Nichol. 2007. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology 36210-15. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

