Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell
- PMID: 18787002
- PMCID: PMC2573255
- DOI: 10.1128/JVI.02375-07
Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell
Abstract
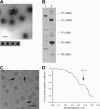
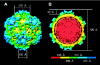
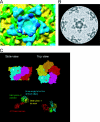
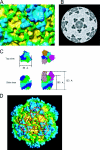
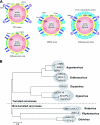
The conservation of the core structure and diversification of the external features among the turreted reoviruses appear to be relevant to structural evolution in facilitating the infection of diverse host species. The structure of Rice ragged stunt virus (RRSV), in the genus Oryzavirus of the family Reoviridae, is determined to show a core composed of capsid shell, clamps, and long turrets. The RRSV core structure is equivalent to the core structure of Orthoreovirus and the virion structure of Cytoplasmic polyhedrosis virus (CPV). In RRSV, five peripheral trimers surround each long turret and sit at the Q trimer position in the T=13l icosahedral symmetry, a structural feature unique to turreted reoviruses. That is, the core of RRSV is partially covered by 60 copies of the peripheral trimer. In contrast, the core of Orthoreovirus is covered by 200 copies of the trimer that sit at the Q, R, S, and T trimer positions. Our results suggest that among the three viruses, RRSV has a structure intermediate between that of Orthoreovirus and the CPV virion. This conclusion coincides with the results of the phylogenetic analysis of amino acid sequences of RNA-dependent RNA polymerases.
Figures


References
-
- Attoui, H., F. Mohd Jaafar, M. Belhouchet, P. Biagini, J. F. Cantaloube, P. de Micco, and X. de Lamballerie. 2005. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus). Virology 343212-223. - PubMed
-
- Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116120-130. - PubMed
-
- Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15655-663. - PubMed
-
- Bellamy, A., and J. Harvey. 1976. Biophysical studies of reovirus type 3. III. A laser light-scattering study of the RNA transcriptase reaction. Virology 7028-36. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

