Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus
- PMID: 18787083
- PMCID: PMC2556262
- DOI: 10.1101/gr.076075.108
Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus
Abstract
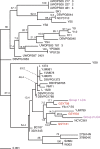
Inter-specific hybridization leading to abrupt speciation is a well-known, common mechanism in angiosperm evolution; only recently, however, have similar hybridization and speciation mechanisms been documented to occur frequently among the closely related group of sensu stricto Saccharomyces yeasts. The economically important lager beer yeast Saccharomyces pastorianus is such a hybrid, formed by the union of Saccharomyces cerevisiae and Saccharomyces bayanus-related yeasts; efforts to understand its complex genome, searching for both biological and brewing-related insights, have been underway since its hybrid nature was first discovered. It had been generally thought that a single hybridization event resulted in a unique S. pastorianus species, but it has been recently postulated that there have been two or more hybridization events. Here, we show that there may have been two independent origins of S. pastorianus strains, and that each independent group--defined by characteristic genome rearrangements, copy number variations, ploidy differences, and DNA sequence polymorphisms--is correlated with specific breweries and/or geographic locations. Finally, by reconstructing common ancestral genomes via array-CGH data analysis and by comparing representative DNA sequences of the S. pastorianus strains with those of many different S. cerevisiae isolates, we have determined that the most likely S. cerevisiae ancestral parent for each of the independent S. pastorianus groups was an ale yeast, with different, but closely related ale strains contributing to each group's parentage.
Figures


References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Awad I.A., Rees C.A., Hernandez-Boussard T., Ball C.A., Sherlock G., Rees C.A., Hernandez-Boussard T., Ball C.A., Sherlock G., Hernandez-Boussard T., Ball C.A., Sherlock G., Ball C.A., Sherlock G., Sherlock G. Caryoscope: An Open Source Java application for viewing microarray data in a genomic context. BMC Bioinformatics. 2004;5:151. doi: 10.1186/1471-2105-5-151. - DOI - PMC - PubMed
-
- Bond U., Neal C., Donnelly D., James T.C., Neal C., Donnelly D., James T.C., Donnelly D., James T.C., James T.C. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr. Genet. 2004;45:360–370. - PubMed
-
- Bozdech Z., Zhu J., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Zhu J., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Cohen F.E., Pulliam B., DeRisi J.L., Pulliam B., DeRisi J.L., DeRisi J.L. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. - DOI - PMC - PubMed
-
- Byrnes J.K., Morris G.P., Li W.H., Morris G.P., Li W.H., Li W.H. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 2006;23:1136–1143. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
