Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes
- PMID: 18787690
- PMCID: PMC2522273
- DOI: 10.1371/journal.ppat.1000144
Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes
Abstract
Streptolysin S (SLS) is a bacteriocin-like haemolytic and cytotoxic virulence factor that plays a key role in the virulence of Group A Streptococcus (GAS), the causative agent of pharyngitis, impetigo, necrotizing fasciitis and streptococcal toxic shock syndrome. Although it has long been thought that SLS and related peptides are produced by GAS and related streptococci only, there is evidence to suggest that a number of the most notorious Gram-positive pathogenic bacteria, including Listeria monocytogenes, Clostridium botulinum and Staphylococcus aureus, produce related peptides. The distribution of the L. monocytogenes cluster is particularly noteworthy in that it is found exclusively among a subset of lineage I strains; i.e., those responsible for the majority of outbreaks of listeriosis. Expression of these genes results in the production of a haemolytic and cytotoxic factor, designated Listeriolysin S, which contributes to virulence of the pathogen as assessed by murine- and human polymorphonuclear neutrophil-based studies. Thus, in the process of establishing the existence of an extended family of SLS-like modified virulence peptides (MVPs), the genetic basis for the enhanced virulence of a proportion of lineage I L. monocytogenes may have been revealed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



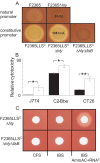
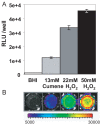
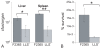
References
-
- Xie L, van der Donk WA. Post-translational modifications during lantibiotic biosynthesis. Curr Opin Chem Biol. 2004;8:498–507. - PubMed
-
- Jack RW, Jung G. Lantibiotics and microcins: polypeptides with unusual chemical diversity. Curr Opin Chem Biol. 2000;4:310–317. - PubMed
-
- Cotter PD, Hill C, Ross RP. Food Microbiology: Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. - PubMed
-
- Cox CR, Coburn PS, Gilmore MS. Enterococcal Cytolysin: A Novel Two Component Peptide System that Serves as a Bacterial Defense Against Eukaryotic and Prokaryotic Cells. Curr Protein Pept Sci. 2005;6:77–84. - PubMed
-
- Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, et al. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56:681–695. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

