Identifying the important HIV-1 recombination breakpoints
- PMID: 18787691
- PMCID: PMC2522274
- DOI: 10.1371/journal.pcbi.1000178
Identifying the important HIV-1 recombination breakpoints
Abstract
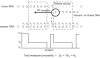
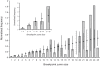
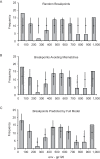
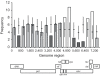
Recombinant HIV-1 genomes contribute significantly to the diversity of variants within the HIV/AIDS pandemic. It is assumed that some of these mosaic genomes may have novel properties that have led to their prevalence, particularly in the case of the circulating recombinant forms (CRFs). In regions of the HIV-1 genome where recombination has a tendency to convey a selective advantage to the virus, we predict that the distribution of breakpoints--the identifiable boundaries that delimit the mosaic structure--will deviate from the underlying null distribution. To test this hypothesis, we generate a probabilistic model of HIV-1 copy-choice recombination and compare the predicted breakpoint distribution to the distribution from the HIV/AIDS pandemic. Across much of the HIV-1 genome, we find that the observed frequencies of inter-subtype recombination are predicted accurately by our model. This observation strongly indicates that in these regions a probabilistic model, dependent on local sequence identity, is sufficient to explain breakpoint locations. In regions where there is a significant over- (either side of the env gene) or under- (short regions within gag, pol, and most of env) representation of breakpoints, we infer natural selection to be influencing the recombination pattern. The paucity of recombination breakpoints within most of the envelope gene indicates that recombinants generated in this region are less likely to be successful. The breakpoints at a higher frequency than predicted by our model are approximately at either side of env, indicating increased selection for these recombinants as a consequence of this region, or at least part of it, having a tendency to be recombined as an entire unit. Our findings thus provide the first clear indication of the existence of a specific portion of the genome that deviates from a probabilistic null model for recombination. This suggests that, despite the wide diversity of recombinant forms seen in the viral population, only a minority of recombination events appear to be of significance to the evolution of HIV-1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. - PubMed
-
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. - PubMed
-
- Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

