Modeling synthetic lethality
- PMID: 18789146
- PMCID: PMC2592713
- DOI: 10.1186/gb-2008-9-9-r135
Modeling synthetic lethality
Abstract
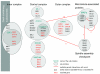
Background: Synthetic lethality defines a genetic interaction where the combination of mutations in two or more genes leads to cell death. The implications of synthetic lethal screens have been discussed in the context of drug development as synthetic lethal pairs could be used to selectively kill cancer cells, but leave normal cells relatively unharmed. A challenge is to assess genome-wide experimental data and integrate the results to better understand the underlying biological processes. We propose statistical and computational tools that can be used to find relationships between synthetic lethality and cellular organizational units.
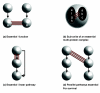
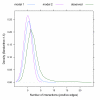
Results: In Saccharomyces cerevisiae, we identified multi-protein complexes and pairs of multi-protein complexes that share an unusually high number of synthetic genetic interactions. As previously predicted, we found that synthetic lethality can arise from subunits of an essential multi-protein complex or between pairs of multi-protein complexes. Finally, using multi-protein complexes allowed us to take into account the pleiotropic nature of the gene products.
Conclusions: Modeling synthetic lethality using current estimates of the yeast interactome is an efficient approach to disentangle some of the complex molecular interactions that drive a cell. Our model in conjunction with applied statistical methods and computational methods provides new tools to better characterize synthetic genetic interactions.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

