A DExH/D-box protein coordinates the two steps of splicing in a group I intron
- PMID: 18789947
- PMCID: PMC2643062
- DOI: 10.1016/j.jmb.2008.08.070
A DExH/D-box protein coordinates the two steps of splicing in a group I intron
Abstract
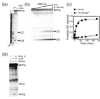
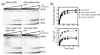
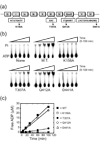
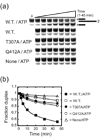
Proteins of the DExH/D family are ATPases that can unwind duplex RNA in vitro. Individual members of this family coordinate many steps in ribonucleoprotein enzyme assembly and catalysis in vivo, but it is largely unknown how the action of these co-factors is specified and precisely timed. As a first step to address this question biochemically, we describe the development of a new protein-dependent group I intron splicing system that requires such an ATPase for coordinating successive steps in splicing. While genetic analysis in yeast has shown that at least five nuclear-encoded proteins are required for splicing of the mitochondrial aI5beta group I intron, we show that efficient in vitro splicing of aI5beta occurs with only two of these co-factors and, furthermore, they fulfill distinct functions in vitro. The Mrs1p protein stabilizes RNA structure and promotes the first step in splicing. In contrast, a DExH/D protein, Mss116p, acts after the first step and, utilizing ATP hydrolysis, specifically enhances the efficiency of exon ligation. An analysis of Mss116p variants with mutations that impair its RNA-stimulated ATP hydrolysis activity or reduce its ability to unwind duplexes show that the efficiency of ATP hydrolysis is a major determinant in promoting exon ligation. These observations suggest that Mss116p acts in aI5beta splicing by catalyzing changes in the structure of the RNA/protein splicing intermediate that promote the second step. More broadly, these observations are consistent with a model in which the "functional-timing" of DExH/D-box protein action can be specified by a specific conformation of its substrate due to the "upstream" activity of other co-factors.
Figures



Similar articles
-
Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP).J Biol Chem. 2010 Mar 19;285(12):8585-94. doi: 10.1074/jbc.M109.090761. Epub 2010 Jan 11. J Biol Chem. 2010. PMID: 20064926 Free PMC article.
-
ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo.J Mol Biol. 2011 Aug 19;411(3):661-79. doi: 10.1016/j.jmb.2011.05.047. Epub 2011 Jun 7. J Mol Biol. 2011. PMID: 21679717 Free PMC article.
-
Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity.J Mol Biol. 2007 Jan 19;365(3):835-55. doi: 10.1016/j.jmb.2006.09.083. Epub 2006 Oct 3. J Mol Biol. 2007. PMID: 17081564 Free PMC article.
-
A new twist on RNA helicases: DExH/D box proteins as RNPases.Nat Struct Biol. 2001 Feb;8(2):113-6. doi: 10.1038/84091. Nat Struct Biol. 2001. PMID: 11175897 Review.
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
Cited by
-
Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP).J Biol Chem. 2010 Mar 19;285(12):8585-94. doi: 10.1074/jbc.M109.090761. Epub 2010 Jan 11. J Biol Chem. 2010. PMID: 20064926 Free PMC article.
-
RNA helicase proteins as chaperones and remodelers.Annu Rev Biochem. 2014;83:697-725. doi: 10.1146/annurev-biochem-060713-035546. Epub 2014 Mar 12. Annu Rev Biochem. 2014. PMID: 24635478 Free PMC article. Review.
-
Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins.J Mol Biol. 2011 Jun 10;409(3):399-414. doi: 10.1016/j.jmb.2011.04.004. Epub 2011 Apr 9. J Mol Biol. 2011. PMID: 21501623 Free PMC article.
-
Mitochondrial RNA Helicases: Key Players in the Regulation of Plant Organellar RNA Splicing and Gene Expression.Int J Mol Sci. 2024 May 17;25(10):5502. doi: 10.3390/ijms25105502. Int J Mol Sci. 2024. PMID: 38791540 Free PMC article. Review.
-
Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing.J Mol Biol. 2010 May 7;398(3):429-43. doi: 10.1016/j.jmb.2010.03.025. Epub 2010 Mar 20. J Mol Biol. 2010. PMID: 20307546 Free PMC article.
References
-
- Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7:831–833. - PubMed
-
- Schroeder R, Grossberger R, Pichler A, Waldsich C. RNA folding in vivo. Curr Opin Struct Biol. 2002;12:296–300. - PubMed
-
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

