Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models
- PMID: 18790843
- PMCID: PMC2586579
- DOI: 10.1529/biophysj.108.139428
Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models
Abstract
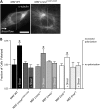
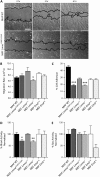
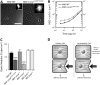
Laminopathies encompass a wide array of human diseases associated to scattered mutations along LMNA, a single gene encoding A-type lamins. How such genetic alterations translate to cellular defects and generate such diverse disease phenotypes remains enigmatic. Recent work has identified nuclear envelope proteins--emerin and the linker of the nucleoskeleton and cytoskeleton (LINC) complex--which connect the nuclear lamina to the cytoskeleton. Here we quantitatively examine the composition of the nuclear envelope, as well as the architecture and functions of the cytoskeleton in cells derived from two laminopathic mouse models, including Hutchinson-Gilford progeria syndrome (Lmna(L530P/L530P)) and Emery-Dreifuss muscular dystrophy (Lmna(-/-)). Cells derived from the overtly aphenotypical model of X-linked Emery-Dreifuss muscular dystrophy (Emd(-/y)) were also included. We find that the centrosome is detached from the nucleus, preventing centrosome polarization in cells under flow--defects that are mediated by the loss of emerin from the nuclear envelope. Moreover, while basal actin and focal adhesion structure are mildly affected, RhoA activation, cell-substratum adhesion, and cytoplasmic elasticity are greatly lowered, exclusively in laminopathic models in which the LINC complex is disrupted. These results indicate a new function for emerin in cell polarization and suggest that laminopathies are not directly associated with cells' inability to polarize, but rather with cytoplasmic softening and weakened adhesion mediated by the disruption of the LINC complex across the nuclear envelope.
Figures




References
-
- Capell, B. C., and F. S. Collins. 2006. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 7:940–952. - PubMed
-
- Eriksson, M., W. T. Brown, L. B. Gordon, M. W. Glynn, J. Singer, L. Scott, M. R. Erdos, C. M. Robbins, T. Y. Moses, P. Berglund, A. Dutra, E. Pak, S. Durkin, A. B. Csoka, M. Boehnke, T. W. Glover, and F. S. Collins. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423:293–298. - PMC - PubMed
-
- Chen, L., L. Lee, B. A. Kudlow, H. G. Dos Santos, O. Sletvold, Y. Shafeghati, E. G. Botha, A. Garg, N. B. Hanson, G. M. Martin, I. S. Mian, B. K. Kennedy, and J. Oshima. 2003. LMNA mutations in atypical Werner's syndrome. Lancet. 362:440–445. - PubMed
-
- Manilal, S., T. M. Nguyen, C. A. Sewry, and G. E. Morris. 1996. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 5:801–808. - PubMed
-
- Charniot, J. C., C. Pascal, C. Bouchier, P. Sebillon, J. Salama, L. Duboscq-Bidot, M. Peuchmaurd, M. Desnos, J. Y. Artigou, and M. Komajda. 2003. Functional consequences of an LMNA mutation associated with a new cardiac and non-cardiac phenotype. Hum. Mutat. 21:473–481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

