Hypernegative supercoiling inhibits growth by causing RNA degradation
- PMID: 18790862
- PMCID: PMC2576660
- DOI: 10.1128/JB.00680-08
Hypernegative supercoiling inhibits growth by causing RNA degradation
Abstract
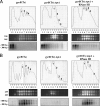
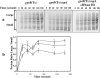
Transcription-induced hypernegative supercoiling is a hallmark of Escherichia coli topoisomerase I (topA) mutants. However, its physiological significance has remained unclear. Temperature downshift of a mutant yielded transient growth arrest and a parallel increase in hypernegative supercoiling that was more severe with lower temperature. Both properties were alleviated by overexpression of RNase HI. While ribosomes in extracts showed normal activity when obtained during growth arrest, mRNA on ribosomes was reduced for fis and shorter for crp, polysomes were much less abundant relative to monosomes, and protein synthesis rate dropped, as did the ratio of large to small proteins. Altered processing and degradation of lacA and fis mRNA was also observed. These data are consistent with truncation of mRNA during growth arrest. These effects were not affected by a mutation in the gene encoding RNase E, indicating that this endonuclease is not involved in the abnormal mRNA processing. They were also unaffected by spectinomycin, an inhibitor of protein synthesis, which argued against induction of RNase activity. In vitro transcription revealed that R-loop formation is more extensive on hypernegatively supercoiled templates. These results allow us, for the first time, to present a model by which hypernegative supercoiling inhibits growth. In this model, the introduction of hypernegative supercoiling by gyrase facilitates degradation of nascent RNA; overproduction of RNase HI limits the accumulation of hypernegative supercoiling, thereby preventing extensive RNA degradation.
Figures






References
-
- Adhya, S., and M. Gottesman. 1978. Control of transcription termination. Annu. Rev. Biochem. 47967-996. - PubMed
-
- Baaklini, I., C. Hraiky, F. Rallu, Y.-C. Tse-Dinh, and M. Drolet. 2004. RNase HI is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 54198-211. - PubMed
-
- Broccoli, S., F. Rallu, P. Sanscartier, S. M. Cerritelli, R. J. Crouch, and M. Drolet. 2004. Effects of RNA polymerase modifications on transcription-induced supercoiling and associated R-loop formation. Mol. Microbiol. 521769-1779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

