Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting
- PMID: 18791027
- PMCID: PMC2576696
- DOI: 10.1128/AEM.00985-08
Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting
Abstract
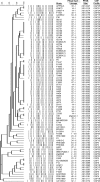
In this study, variably absent or present (VAP) regions discovered through comparative genomics experiments were targeted for the development of a rapid, PCR-based method to subtype and fingerprint Escherichia coli O157:H7. Forty-four VAP loci were analyzed for discriminatory power among 79 E. coli O157:H7 strains of 13 phage types (PT). Twenty-three loci were found to maximize resolution among strains, generating 54 separate fingerprints, each of which contained strains of unique PT. Strains from the three previously identified major E. coli O157:H7 lineages, LSPA6-LI, LSPA6-LI/II, and LSPA6-LII, formed distinct branches on a dendrogram obtained by hierarchical clustering of comparative genomic fingerprinting (CGF) data. By contrast, pulsed-field gel electrophoresis (PFGE) typing generated 52 XbaI digestion profiles that were not unique to PT and did not cluster according to O157:H7 lineage. Our analysis identified a subpopulation comprised of 25 strains from a closed herd of cattle, all of which were of PT87 and formed a cluster distinct from all other E. coli O157:H7 strains examined. CGF found five related but unique fingerprints among the highly clonal herd strains, with two dominant subtypes characterized by a shift from the presence of locus fprn33 to its absence. CGF had equal resolution to PFGE typing but with greater specificity, generating fingerprints that were unique among phenotypically related E. coli O157:H7 lineages and PT. As a comparative genomics typing method that is amenable for use in high-throughput platforms, CGF may be a valuable tool in outbreak investigations and strain characterization.
Figures

References
-
- Ahmed, R., C. Bopp, A. Borczyk, and S. Kasatiya. 1987. Phage-typing scheme for Escherichia coli O157:H7. J. Infect. Dis. 155:806-809. - PubMed
-
- Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. - PMC - PubMed
-
- Avery, S. M., E. Liebana, C. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. - PMC - PubMed
-
- Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. - PMC - PubMed
-
- Beutin, L., S. Kaulfuss, S. Herold, E. Oswald, and H. Schmidt. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552-1563. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

