Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements
- PMID: 18791231
- PMCID: PMC2581937
- DOI: 10.1534/genetics.108.093211
Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements
Abstract
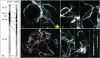
Ongoing genomics projects of tomato (Solanum lycopersicum) and potato (S. tuberosum) are providing unique tools for comparative mapping studies in Solanaceae. At the chromosomal level, bacterial artificial chromosomes (BACs) can be positioned on pachytene complements by fluorescence in situ hybridization (FISH) on homeologous chromosomes of related species. Here we present results of such a cross-species multicolor cytogenetic mapping of tomato BACs on potato chromosomes 6 and vice versa. The experiments were performed under low hybridization stringency, while blocking with Cot-100 was essential in suppressing excessive hybridization of repeat signals in both within-species FISH and cross-species FISH of tomato BACs. In the short arm we detected a large paracentric inversion that covers the whole euchromatin part with breakpoints close to the telomeric heterochromatin and at the border of the short arm pericentromere. The long arm BACs revealed no deviation in the colinearity between tomato and potato. Further comparison between tomato cultivars Cherry VFNT and Heinz 1706 revealed colinearity of the tested tomato BACs, whereas one of the six potato clones (RH98-856-18) showed minor putative rearrangements within the inversion. Our results present cross-species multicolor BAC-FISH as a unique tool for comparative genetic studies across Solanum species.
Figures


References
-
- Ammiraju, J. S. S., J. C. Veremis, X. Huang, P. A. Roberts and I. Kaloshian, 2003. The heat-stable root-nematode resistance gene Mi-9 from Lycopersicon peruvianum is localized on the short arm of chromosome 6. Theor. Appl. Genet. 106 478–484. - PubMed
-
- Bai, Y., R. van der Hulst, C. C. Huang, L. Wei, P. Stam et al., 2004. Mapping Ol-4, a gene conferring resistance to Oidium neolycopersici and originating from Lycopersicon peruvianum LA2172, requires multi-allelic, single-locus markers. Theor. Appl. Genet. 109 1215–1223. - PubMed
-
- Blackburn, E. H., 1991. Structure and function of telomeres. Nature 350 569–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

