Controlling type-I error of the McDonald-Kreitman test in genomewide scans for selection on noncoding DNA
- PMID: 18791238
- PMCID: PMC2581974
- DOI: 10.1534/genetics.108.091850
Controlling type-I error of the McDonald-Kreitman test in genomewide scans for selection on noncoding DNA
Abstract
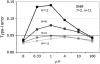
Departures from the assumption of homogenously interdigitated neutral and putatively selected sites in the McDonald-Kreitman test can lead to false rejections of the neutral model in the presence of intermediate levels of recombination. This problem is exacerbated by small sample sizes, nonequilibrium demography, recombination rate variation, and in comparisons involving more recently diverged species. I propose that establishing significance levels by coalescent simulation with recombination can improve the fidelity of the test in genomewide scans for selection on noncoding DNA.
Figures


References
-
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437 1149–1152. - PubMed
-
- Bachtrog, D., K. Thornton, A. Clark and P. Andolfatto, 2006. Extensive introgression of mitochondrial DNA relative to nuclear gene flow in the Drosophila yakuba species group. Evol. Int. J. Org. Evol. 60 292–302. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

