Transcriptional autoregulatory loops are highly conserved in vertebrate evolution
- PMID: 18791639
- PMCID: PMC2527657
- DOI: 10.1371/journal.pone.0003210
Transcriptional autoregulatory loops are highly conserved in vertebrate evolution
Abstract
Background: Feedback loops are the simplest building blocks of transcriptional regulatory networks and therefore their behavior in the course of evolution is of prime interest.
Methodology: We address the question of enrichment of the number of autoregulatory feedback loops in higher organisms. First, based on predicted autoregulatory binding sites we count the number of autoregulatory loops. We compare it to estimates obtained either by assuming that each (conserved) gene has the same chance to be a target of a given factor or by assuming that each conserved sequence position has an equal chance to be a binding site of the factor.
Conclusions: We demonstrate that the numbers of putative autoregulatory loops conserved between human and fugu, danio or chicken are significantly higher than expected. Moreover we show, that conserved autoregulatory binding sites cluster close to the factors' starts of transcription. We conclude, that transcriptional autoregulatory feedback loops constitute a core transcriptional network motif and their conservation has been maintained in higher vertebrate organism evolution.
Conflict of interest statement
Figures

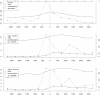
 predicted to have a conserved binding site of f. Vertical axes give the fraction of regulated genes
predicted to have a conserved binding site of f. Vertical axes give the fraction of regulated genes  . Three cases are shown: no conservation, conservation to danio and to fugu.
. Three cases are shown: no conservation, conservation to danio and to fugu.
Similar articles
-
ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes.Bioinformatics. 2007 Jan 1;23(1):122-4. doi: 10.1093/bioinformatics/btl546. Epub 2006 Nov 7. Bioinformatics. 2007. PMID: 17090579
-
Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster.Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3819-24. doi: 10.1073/pnas.0800398105. Epub 2008 Mar 4. Proc Natl Acad Sci U S A. 2008. PMID: 18319338 Free PMC article.
-
Characterization of chicken octamer-binding proteins demonstrates that POU domain-containing homeobox transcription factors have been highly conserved during vertebrate evolution.Proc Natl Acad Sci U S A. 1990 Feb;87(3):1099-103. doi: 10.1073/pnas.87.3.1099. Proc Natl Acad Sci U S A. 1990. PMID: 1967834 Free PMC article.
-
[The role of conserved sequences in the regulatory elements of the Antp-like homeobox-containing genes of vertebrates].Zh Evol Biokhim Fiziol. 1996 Sep-Oct;32(5):556-68. Zh Evol Biokhim Fiziol. 1996. PMID: 9092235 Review. Russian.
-
Comparative genomics using fugu: a tool for the identification of conserved vertebrate cis-regulatory elements.Bioessays. 2005 Jan;27(1):100-7. doi: 10.1002/bies.20134. Bioessays. 2005. PMID: 15612032 Review.
Cited by
-
Stochastic signalling rewires the interaction map of a multiple feedback network during yeast evolution.Nat Commun. 2012 Feb 21;3:682. doi: 10.1038/ncomms1687. Nat Commun. 2012. PMID: 22353713 Free PMC article.
-
Making sense of transcription networks.Cell. 2015 May 7;161(4):714-23. doi: 10.1016/j.cell.2015.04.014. Cell. 2015. PMID: 25957680 Free PMC article. Review.
-
Dynamical gene regulatory networks are tuned by transcriptional autoregulation with microRNA feedback.Sci Rep. 2020 Jul 31;10(1):12960. doi: 10.1038/s41598-020-69791-5. Sci Rep. 2020. PMID: 32737375 Free PMC article.
-
TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop.Nat Cell Biol. 2013 Jun;15(6):647-58. doi: 10.1038/ncb2718. Epub 2013 Apr 21. Nat Cell Biol. 2013. PMID: 23604321 Free PMC article.
-
Using evolutionary conserved modules in gene networks as a strategy to leverage high throughput gene expression queries.PLoS One. 2010 Sep 2;5(9):e12525. doi: 10.1371/journal.pone.0012525. PLoS One. 2010. PMID: 20824082 Free PMC article.
References
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–61. - PubMed
-
- Tao Y. Intrinsic and external noise in an auto-regulatory genetic network. J Theor Biol. 2004;229(2):147–56. - PubMed
-
- Bai G, Sheng N, Xie Z, Bian W, Yokota Y, et al. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13(2):283–97. - PubMed
-
- Smith S, Watada H, Scheel D, Mrejen C, German M. Autoregulation and maturity onset diabetes of the young transcription factors control the human PAX4 promoter. J Biol Chem. 2000;275(47):36910–9. - PubMed
-
- Johnson D, Ohtani K, Nevins J. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8(13):1514–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

