Interactions between M protein and other structural proteins of severe, acute respiratory syndrome-associated coronavirus
- PMID: 18792806
- PMCID: PMC7089546
- DOI: 10.1007/s11373-008-9278-3
Interactions between M protein and other structural proteins of severe, acute respiratory syndrome-associated coronavirus
Abstract
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) structural proteins (S, E, M, and NC) localize in different subcellular positions when expressed individually. However, SARS-CoV M protein is co-localized almost entirely with S, E, or NC protein when co-expressed in the cells. On the other hand, only partial co-localization was observed when S and E, S and NC, or E and NC were co-expressed in the cells. Interactions between SARS-CoV M and other structural proteins but not interactions between S and E, S and NC, or E and NC were further demonstrated by co-immunoprecipitation assay. These results indicate that SARS-CoV M protein, similar to the M proteins of other coronaviruses, plays a pivotal role in virus assembly. The cytoplasmic C-terminus domain of SARS-CoV M protein was responsible for binding to NC protein. Multiple regions of M protein interacted with E and S proteins. A model for the interactions between SARS-CoV M protein and other structural proteins is proposed. This study helps us better understand protein-protein interactions during viral assembly of SARS-CoV.
Figures
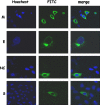
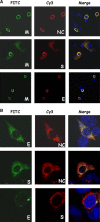


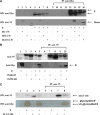
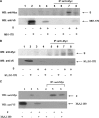
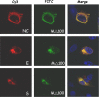
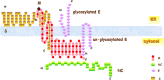
References
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. - DOI - PubMed
-
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stohr K, Peiris JS, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. - DOI - PMC - PubMed
-
- Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
