Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011
- PMID: 18793445
- PMCID: PMC2573895
- DOI: 10.1186/1471-2164-9-416
Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011
Abstract
Background: Small non-coding RNAs (sRNAs) have emerged as ubiquitous regulatory elements in bacteria and other life domains. However, few sRNAs have been identified outside several well-studied species of gamma-proteobacteria and thus relatively little is known about the role of RNA-mediated regulation in most other bacterial genera. Here we have conducted a computational prediction of putative sRNA genes in intergenic regions (IgRs) of the symbiotic alpha-proteobacterium S. meliloti 1021 and experimentally confirmed the expression of dozens of these candidate loci in the closely related strain S. meliloti 2011.
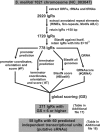
Results: Our first sRNA candidate compilation was based mainly on the output of the sRNAPredictHT algorithm. A thorough manual sequence analysis of the curated list rendered an initial set of 18 IgRs of interest, from which 14 candidates were detected in strain 2011 by Northern blot and/or microarray analysis. Interestingly, the intracellular transcript levels varied in response to various stress conditions. We developed an alternative computational method to more sensitively predict sRNA-encoding genes and score these predicted genes based on several features to allow identification of the strongest candidates. With this novel strategy, we predicted 60 chromosomal independent transcriptional units that, according to our annotation, represent strong candidates for sRNA-encoding genes, including most of the sRNAs experimentally verified in this work and in two other contemporary studies. Additionally, we predicted numerous candidate sRNA genes encoded in megaplasmids pSymA and pSymB. A significant proportion of the chromosomal- and megaplasmid-borne putative sRNA genes were validated by microarray analysis in strain 2011.
Conclusion: Our data extend the number of experimentally detected S. meliloti sRNAs and significantly expand the list of putative sRNA-encoding IgRs in this and closely related alpha-proteobacteria. In addition, we have developed a computational method that proved useful to predict sRNA-encoding genes in S. meliloti. We anticipate that this predictive approach can be flexibly implemented in many other bacterial species.
Figures


References
-
- Storz G, Haas D. A guide to small RNAs in microorganisms. Curr Opin Microbiol. 2007;10:93–95. doi: 10.1016/j.mib.2007.03.017. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

