Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts
- PMID: 18794137
- PMCID: PMC2556890
- DOI: 10.1158/0008-5472.CAN-08-1404
Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts
Abstract
The effectiveness of therapies targeting specific pathways in breast cancer, such as the estrogen receptor or HER2, is limited because many tumors manifest resistance, either de novo or acquired, during the course of treatment. To investigate molecular mechanisms of resistance, we used two xenograft models of estrogen receptor-positive (ER+) breast cancer, one with and one without HER2 overexpression (MCF7/HER2-18 and MCF7 wt, respectively). Mice with established tumors were assigned to the following treatment groups: estrogen supplementation (E2), estrogen deprivation (ED), ED plus tamoxifen (Tam), all with or without the epidermal growth factor receptor tyrosine kinase inhibitor gefitinib (G). Another group received ED plus the antiestrogen fulvestrant (MCF7 wt only). Tumors with acquired or de novo resistance to these endocrine therapies were profiled for gene expression and compared with tumors in the E2 control group. One class of genes underexpressed in endocrine-resistant tumors (relative to E2-treated tumors) were estrogen inducible in vitro and associated with ER+ human breast cancers (luminal subtype). Another class of genes overexpressed in tumors with acquired resistance in both models represented transcriptional targets of HER2 signaling and was associated with ER-/HER2+ human cancers (ERBB2+ subtype). A third class of genes overexpressed in MCF7/HER2-18 tumors exhibiting de novo resistance to tamoxifen was associated with ER+ human cancers but not with estrogen-regulated genes. Thus, in response to various endocrine therapy regimens, these xenograft breast tumors shut down classic estrogen signaling and activate alternative pathways such as HER2 that contribute to treatment resistance. Over time, the molecular phenotype of breast cancer can change.
Conflict of interest statement
Figures
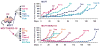



References
-
- Clark GM, McGuire WL. Steroid receptors and other prognostic factors in primary breast cancer. Semin Oncol. 1988;15(2 Suppl 1):20–5. - PubMed
-
- Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–12. - PubMed
-
- Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339(22):1609–18. - PubMed
-
- Schiff R, Reddy P, Ahotupa M, et al. Oxidative stress and AP-1 activity in tamoxifen-resistant breast tumors in vivo. J Natl Cancer Inst. 2000;92(23):1926–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

