Regulation of Sli15/INCENP, kinetochore, and Cdc14 phosphatase functions by the ribosome biogenesis protein Utp7
- PMID: 18794331
- PMCID: PMC2542472
- DOI: 10.1083/jcb.200802085
Regulation of Sli15/INCENP, kinetochore, and Cdc14 phosphatase functions by the ribosome biogenesis protein Utp7
Abstract
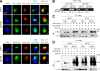
The Sli15-Ipl1-Bir1 chromosomal passenger complex is essential for proper kinetochore-microtubule attachment and spindle stability in the budding yeast Saccharomyces cerevisiae. During early anaphase, release of the Cdc14 protein phosphatase from the nucleolus leads to the dephosphorylation of Sli15 and redistribution of this complex from kinetochores to the spindle. We show here that the predominantly nucleolar ribosome biogenesis protein Utp7 is also present at kinetochores and is required for normal organization of kinetochore proteins and proper chromosome segregation. Utp7 associates with and regulates the localization of Sli15 and Cdc14. Before anaphase onset, it prevents the premature nucleolar release of Cdc14 and the premature concentration of Sli15 on the spindle. Furthermore, Utp7 can regulate the localization and phosphorylation status of Sli15 independent of its effect on Cdc14 function. Thus, Utp7 is a multifunctional protein that plays essential roles in the vital cellular processes of ribosome biogenesis, chromosome segregation, and cell cycle control.
Figures
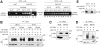
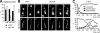

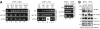


References
-
- Andersen, J.S., C.E. Lyon, A.H. Fox, A.K.L. Leung, Y.W. Lam, H. Steen, M. Mann, and A.I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1–11. - PubMed
-
- Andersen, J.S., Y.W. Lam, A.K.L. Leung, S.-E. Ong, C.E. Lyon, A.I. Lamond, and M. Mann. 2005. Nucleolar proteome dynamics. Nature. 433:77–83. - PubMed
-
- Azzam, R., S.L. Chen, W. Shou, A.S. Mah, G. Alexandru, K. Nasmyth, R.S. Annan, S.A. Carr, and R.J. Deshaies. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 305:516–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

