Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae
- PMID: 18794362
- PMCID: PMC2573294
- DOI: 10.1128/MCB.01210-08
Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae
Abstract
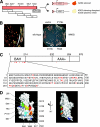
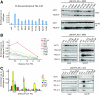
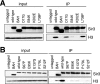
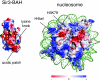
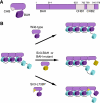
Silent chromatin in Saccharomyces cerevisiae is established in a stepwise process involving the SIR complex, comprised of the histone deacetylase Sir2 and the structural components Sir3 and Sir4. The Sir3 protein, which is the primary histone-binding component of the SIR complex, forms oligomers in vitro and has been proposed to mediate the spreading of the SIR complex along the chromatin fiber. In order to analyze the role of Sir3 in the spreading of the SIR complex, we performed a targeted genetic screen for alleles of SIR3 that dominantly disrupt silencing. Most mutations mapped to a single surface in the conserved N-terminal BAH domain, while one, L738P, localized to the AAA ATPase-like domain within the C-terminal half of Sir3. The BAH point mutants, but not the L738P mutant, disrupted the interaction between Sir3 and nucleosomes. In contrast, Sir3-L738P bound the N-terminal tail of histone H4 more strongly than wild-type Sir3, indicating that misregulation of the Sir3 C-terminal histone-binding activity also disrupted spreading. Our results underscore the importance of proper interactions between Sir3 and the nucleosome in silent chromatin assembly. We propose a model for the spreading of the SIR complex along the chromatin fiber through the two distinct histone-binding domains in Sir3.
Figures






References
-
- Ausubel, F. M. 2002. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology, 5th ed. Wiley, New York, NY.
-
- Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi, and B. Stillman. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83563-568. - PubMed
-
- Buker, S. M., T. Iida, M. Bühler, J. Villén, S. P. Gygi, J. Nakayama, and D. Moazed. 2007. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 14200-207. - PubMed
-
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
