Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding
- PMID: 18794367
- PMCID: PMC2573310
- DOI: 10.1128/MCB.01147-08
Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding
Abstract
Selection of the AUG start codon for translation in eukaryotes is governed by codon-anticodon interactions between the initiator Met-tRNA(i)(Met) and the mRNA. Translation initiation factor 2 (eIF2) binds Met-tRNA(i)(Met) to the 40S ribosomal subunit, and previous studies identified Sui(-) mutations in eIF2 that enhanced initiation from a noncanonical UUG codon, presumably by impairing Met-tRNA(i)(Met) binding. Consistently, an eIF2gamma-N135D GTP-binding domain mutation impairs Met-tRNA(i)(Met) binding and causes a Sui(-) phenotype. Intragenic A208V and A382V suppressor mutations restore Met-tRNA(i)(Met) binding affinity and cell growth; however, only A208V suppresses the Sui(-) phenotype associated with the eIF2gamma-N135D mutation. An eIF2gamma-A219T mutation impairs Met-tRNA(i)(Met) binding but unexpectedly enhances the fidelity of initiation, suppressing the Sui(-) phenotype associated with the eIF2gamma-N135D,A382V mutant. Overexpression of eIF1, which is thought to monitor codon-anticodon interactions during translation initiation, likewise suppresses the Sui(-) phenotype of the eIF2gamma mutants. We propose that structural alterations in eIF2gamma subtly alter the conformation of Met-tRNA(i)(Met) on the 40S subunit and thereby affect the fidelity of start codon recognition independent of Met-tRNA(i)(Met) binding affinity.
Figures
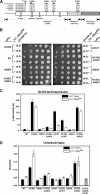
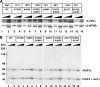
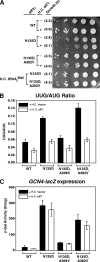

References
-
- Algire, M. A., D. Maag, and J. R. Lorsch. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20251-262. - PubMed
-
- Alone, P. V., and T. E. Dever. 2006. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2Bɛ. J. Biol. Chem. 28112636-12644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
