Activation of tyrosine kinases by mutation of the gatekeeper threonine
- PMID: 18794843
- PMCID: PMC2575426
- DOI: 10.1038/nsmb.1486
Activation of tyrosine kinases by mutation of the gatekeeper threonine
Abstract
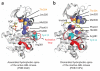
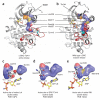
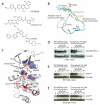
Protein kinases targeted by small-molecule inhibitors develop resistance through mutation of the 'gatekeeper' threonine residue of the active site. Here we show that the gatekeeper mutation in the cellular forms of c-ABL, c-SRC, platelet-derived growth factor receptor-alpha and -beta, and epidermal growth factor receptor activates the kinase and promotes malignant transformation of BaF3 cells. Structural analysis reveals that a network of hydrophobic interactions-the hydrophobic spine-characteristic of the active kinase conformation is stabilized by the gatekeeper substitution. Substitution of glycine for the residues constituting the spine disrupts the hydrophobic connectivity and inactivates the kinase. Furthermore, a small-molecule inhibitor that maximizes complementarity with the dismantled spine (compound 14) inhibits the gatekeeper mutation of BCR-ABL-T315I. These results demonstrate that mutation of the gatekeeper threonine is a common mechanism of activation for tyrosine kinases and provide structural insights to guide the development of next-generation inhibitors.
Figures



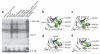
References
-
- Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. - PubMed
-
- Dibb NJ, Dilworth SM, Mol CD. Switching on kinases: oncogenic activation of BRAF and the PDGFR family. Nat. Rev. Cancer. 2004;4:718–727. - PubMed
-
- Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. - PubMed
-
- Sawyers CL. Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genes Dev. 2003;17:2998–3010. - PubMed
-
- Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

