Worlds within worlds: evolution of the vertebrate gut microbiota
- PMID: 18794915
- PMCID: PMC2664199
- DOI: 10.1038/nrmicro1978
Worlds within worlds: evolution of the vertebrate gut microbiota
Abstract
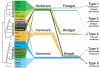
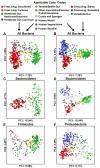
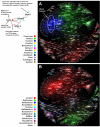
In this Analysis we use published 16S ribosomal RNA gene sequences to compare the bacterial assemblages that are associated with humans and other mammals, metazoa and free-living microbial communities that span a range of environments. The composition of the vertebrate gut microbiota is influenced by diet, host morphology and phylogeny, and in this respect the human gut bacterial community is typical of an omnivorous primate. However, the vertebrate gut microbiota is different from free-living communities that are not associated with animal body habitats. We propose that the recently initiated international Human Microbiome Project should strive to include a broad representation of humans, as well as other mammalian and environmental samples, as comparative analyses of microbiotas and their microbiomes are a powerful way to explore the evolutionary history of the biosphere.
Figures


References
-
- Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. - PubMed
-
- Martiny JB, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. - PubMed
-
-
Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U S A. 2007;104:11436–11440. This paper combines data from hundreds of bacterial communities to show that phylogenetic tree-based measures of diversity (i.e.,UniFrac) can reveal large-scale trends influencing these communities: for example, the unexpectedly large effect of salinity.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

