Mechanism of partial agonism at the GluR2 AMPA receptor: Measurements of lobe orientation in solution
- PMID: 18795801
- PMCID: PMC2756675
- DOI: 10.1021/bi800843c
Mechanism of partial agonism at the GluR2 AMPA receptor: Measurements of lobe orientation in solution
Abstract
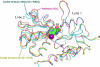
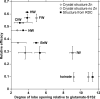
The mechanism by which the binding of a neurotransmitter to a receptor leads to channel opening is a central issue in molecular neurobiology. The structure of the agonist binding domain of ionotropic glutamate receptors has led to an improved understanding of the changes in structure that accompany agonist binding and have provided important clues about the link between these structural changes and channel activation and desensitization. However, because the binding domain has exhibited different structures under different crystallization conditions, understanding the structure in the absence of crystal packing is of considerable importance. The orientation of the two lobes of the binding domain in the presence of a full agonist, an antagonist, and several partial agonists was measured using NMR spectroscopy by employing residual dipolar couplings. For some partial agonists, the solution conformation differs from that observed in the crystal. A model of channel activation based on the results is discussed.
Figures



References
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. - PubMed
-
- Dingledine R, Borges K, Bowie D, Traynelis S. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. - PubMed
-
- Asztely F, Gustafsson B. Ionotropic glutamate receptors. Their possible role in the expression of hippocampal synaptic plasticity. Mol Neurobiol. 1996;12:1–11. - PubMed
-
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

