Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse
- PMID: 18796159
- PMCID: PMC2559853
- DOI: 10.1186/1471-2164-9-419
Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse
Abstract
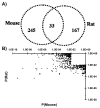
Background: Mouse and rat models are mainstays in pharmacology, toxicology and drug development -- but differences between strains and between species complicate data interpretation and application to human health. Dioxin-like polyhalogenated aromatic hydrocarbons represent a major class of environmentally and economically relevant toxicants. In mammals dioxin exposure leads to a broad spectrum of adverse affects, including hepatotoxicity of varying severity. Several studies have shown that dioxins extensively alter hepatic mRNA levels. Surprisingly, though, analysis of a limited portion of the transcriptome revealed that rat and mouse responses diverge greatly (Boverhof et al. Toxicol Sci 94:398-416, 2006).
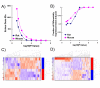
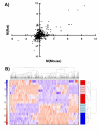
Results: We employed oligonucleotide arrays to compare the response of 8,125 rat and mouse orthologs. We confirmed that there is limited inter-species overlap in dioxin-responsive genes. Rat-specific and mouse-specific genes are enriched for specific functional groups which differ between species, conceivably accounting for species-specificities in liver histopathology. While no evidence for the involvement of copy-number variation was found, extensive inter-species variation in the transcriptional-regulatory network was identified; Nr2f1 and Fos emerged as candidates to explain species-specific and species-independent responses, respectively.
Conclusion: Our results suggest that a small core of genes is responsible for mediating the similar features of dioxin hepatotoxicity in rats and mice but non-overlapping pathways are simultaneously at play to result in distinctive histopathological outcomes. The extreme divergence between mouse and rat transcriptomic responses appears to reflect divergent transcriptional-regulatory networks. Taken together, these data suggest that both rat and mouse models should be used to screen the acute hepatotoxic effects of drugs and toxic compounds.
Figures

References
-
- Pohjanvirta R, Tuomisto J. Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: effects, mechanisms, and animal models. Pharmacol Rev. 1994;46:483–549. - PubMed
-
- Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. - DOI - PubMed

