Characterization of hepatitis C RNA-containing particles from human liver by density and size
- PMID: 18796720
- PMCID: PMC2557069
- DOI: 10.1099/vir.0.2008/000083-0
Characterization of hepatitis C RNA-containing particles from human liver by density and size
Abstract
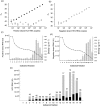
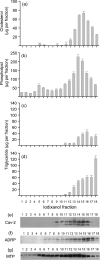
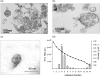
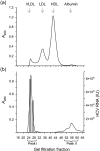
Hepatitis C virus (HCV) particles found in vivo are heterogeneous in density and size, but their detailed characterization has been restricted by the low titre of HCV in human serum. Previously, our group has found that HCV circulates in blood in association with very-low-density lipoprotein (VLDL). Our aim in this study was to characterize HCV RNA-containing membranes and particles in human liver by both density and size and to identify the subcellular compartment(s) where the association with VLDL occurs. HCV was purified by density using iodixanol gradients and by size using gel filtration. Both positive-strand HCV RNA (present in virus particles) and negative-strand HCV RNA (an intermediate in virus replication) were found with densities below 1.08 g ml(-1). Viral structural and non-structural proteins, host proteins ApoB, ApoE and caveolin-2, as well as cholesterol, triglyceride and phospholipids were also detected in these low density fractions. After fractionation by size with Superose gel filtration, HCV RNA and viral proteins co-fractionated with endoplasmic reticulum proteins and VLDL. Fractionation on Toyopearl, which separates particles with diameters up to 200 nm, showed that 78 % of HCV RNA from liver was >100 nm in size, with a positive-/negative-strand ratio of 6 : 1. Also, 8 % of HCV RNA was found in particles with diameters between 40 nm and 70 nm and a positive-/negative-strand ratio of 45 : 1. This HCV was associated with ApoB, ApoE and viral glycoprotein E2, similar to viral particles circulating in serum. Our results indicate that the association between HCV and VLDL occurs in the liver.
Figures

References
-
- Aizaki, H., Lee, K. J., Sung, V. M. H., Ishiko, H. & Lai, M. M. C. (2004). Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324, 450–461. - PubMed
-
- Anonymous (2002). Gel filtration. Principles and Methods. Uppsala, Sweden: Amersham Biosciences.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

