Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover
- PMID: 18799019
- PMCID: PMC2569965
- DOI: 10.1186/1476-4598-7-72
Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover
Abstract
Background: Apart from genetic alterations, development and progression of colorectal cancer has been linked to influences from nutritional intake, hyperalimentation, and cellular metabolic changes that may be the basis for new diagnostic and therapeutic approaches. However, in contrast to genomics and proteomics, comprehensive metabolomic investigations of alterations in malignant tumors have rarely been conducted.
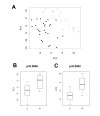
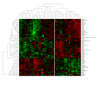
Results: In this study we investigated a set of paired samples of normal colon tissue and colorectal cancer tissue with gas-chromatography time-of-flight mass-spectrometry, which resulted in robust detection of a total of 206 metabolites. Metabolic phenotypes of colon cancer and normal tissues were different at a Bonferroni corrected significance level of p=0.00170 and p=0.00005 for the first two components of an unsupervised PCA analysis. Subsequent supervised analysis found 82 metabolites to be significantly different at p<0.01. Metabolites were connected to abnormalities in metabolic pathways by a new approach that calculates the distance of each pair of metabolites in the KEGG database interaction lattice. Intermediates of the TCA cycle and lipids were found down-regulated in cancer, whereas urea cycle metabolites, purines, pyrimidines and amino acids were generally found at higher levels compared to normal colon mucosa.
Conclusion: This study demonstrates that metabolic profiling facilitates biochemical phenotyping of normal and neoplastic colon tissue at high significance levels and points to GC-TOF-based metabolomics as a new method for molecular pathology investigations.
Figures





References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. - PubMed
-
- Cousins RJ. Nutritional regulation of gene expression. Am J Med. 1999;106:20S–23S. - PubMed
-
- Jump DB. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41–78. - PubMed
-
- Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–72. - PubMed
-
- Gunter MJ, Leitzmann MF. Obesity and colorectal cancer: epidemiology, mechanisms and candidate genes. J Nutr Biochem. 2006;17:145–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

