The Hsk1(Cdc7) replication kinase regulates origin efficiency
- PMID: 18799612
- PMCID: PMC2592646
- DOI: 10.1091/mbc.e08-06-0645
The Hsk1(Cdc7) replication kinase regulates origin efficiency
Abstract
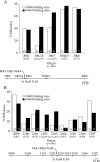
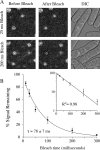
Origins of DNA replication are generally inefficient, with most firing in fewer than half of cell cycles. However, neither the mechanism nor the importance of the regulation of origin efficiency is clear. In fission yeast, origin firing is stochastic, leading us to hypothesize that origin inefficiency and stochasticity are the result of a diffusible, rate-limiting activator. We show that the Hsk1-Dfp1 replication kinase (the fission yeast Cdc7-Dbf4 homologue) plays such a role. Increasing or decreasing Hsk1-Dfp1 levels correspondingly increases or decreases origin efficiency. Furthermore, tethering Hsk1-Dfp1 near an origin increases the efficiency of that origin, suggesting that the effective local concentration of Hsk1-Dfp1 regulates origin firing. Using photobleaching, we show that Hsk1-Dfp1 is freely diffusible in the nucleus. These results support a model in which the accessibility of replication origins to Hsk1-Dfp1 regulates origin efficiency and provides a potential mechanistic link between chromatin structure and replication timing. By manipulating Hsk1-Dfp1 levels, we show that increasing or decreasing origin firing rates leads to an increase in genomic instability, demonstrating the biological importance of appropriate origin efficiency.
Figures


Comment in
- Mol Biol Cell. 19:5031.
References
-
- Bechhoefer J., Marshall B. How Xenopus laevis replicates DNA reliably even though its origins of replication are located and initiated stochastically. Phys. Rev. Lett. 2007;98 098105. - PubMed
-
- Bensimon A., Simon A., Chiffaudel A., Croquette V., Heslot F., Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

