Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46,000 BAC end sequences
- PMID: 18801166
- PMCID: PMC2561056
- DOI: 10.1186/1471-2164-9-423
Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46,000 BAC end sequences
Abstract
Background: Citrus species constitute one of the major tree fruit crops of the subtropical regions with great economic importance. However, their peculiar reproductive characteristics, low genetic diversity and the long-term nature of tree breeding mostly impair citrus variety improvement. In woody plants, genomic science holds promise of improvements and in the Citrus genera the development of genomic tools may be crucial for further crop improvements. In this work we report the characterization of three BAC libraries from Clementine (Citrus clementina), one of the most relevant citrus fresh fruit market cultivars, and the analyses of 46.000 BAC end sequences. Clementine is a diploid plant with an estimated haploid genome size of 367 Mb and 2n = 18 chromosomes, which makes feasible the use of genomics tools to boost genetic improvement.
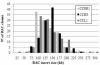
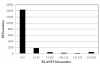
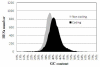
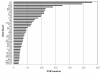
Results: Three genomic BAC libraries of Citrus clementina were constructed through EcoRI, MboI and HindIII digestions and 56,000 clones, representing an estimated genomic coverage of 19.5 haploid genome-equivalents, were picked. BAC end sequencing (BES) of 28,000 clones produced 28.1 Mb of genomic sequence that allowed the identification of the repetitive fraction (12.5% of the genome) and estimation of gene content (31,000 genes) of this species. BES analyses identified 3,800 SSRs and 6,617 putative SNPs. Comparative genomic studies showed that citrus gene homology and microsyntheny with Populus trichocarpa was rather higher than with Arabidopsis thaliana, a species phylogenetically closer to citrus.
Conclusion: In this work, we report the characterization of three BAC libraries from C. clementina, and a new set of genomic resources that may be useful for isolation of genes underlying economically important traits, physical mapping and eventually crop improvement in Citrus species. In addition, BAC end sequencing has provided a first insight on the basic structure and organization of the citrus genome and has yielded valuable molecular markers for genetic mapping and cloning of genes of agricultural interest. Paired end sequences also may be very helpful for whole-genome sequencing programs.
Figures

References
-
- Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet. 2000;100:1155–1166.
-
- Gmitter FG., Jr Origin, evolution and breeding of the grapefruit. Plant Breeding Reviews. 1995;13:345–363.
-
- Forment J, Gadea J, Huerta L, Abizanda L, Agusti J, Alamar S, et al. Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol Biol. 2005;57:375–391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

