MetWAMer: eukaryotic translation initiation site prediction
- PMID: 18801175
- PMCID: PMC2603428
- DOI: 10.1186/1471-2105-9-381
MetWAMer: eukaryotic translation initiation site prediction
Abstract
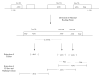
Background: Translation initiation site (TIS) identification is an important aspect of the gene annotation process, requisite for the accurate delineation of protein sequences from transcript data. We have developed the MetWAMer package for TIS prediction in eukaryotic open reading frames of non-viral origin. MetWAMer can be used as a stand-alone, third-party tool for post-processing gene structure annotations generated by external computational programs and/or pipelines, or directly integrated into gene structure prediction software implementations.
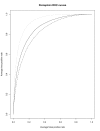
Results: MetWAMer currently implements five distinct methods for TIS prediction, the most accurate of which is a routine that combines weighted, signal-based translation initiation site scores and the contrast in coding potential of sequences flanking TISs using a perceptron. Also, our program implements clustering capabilities through use of the k-medoids algorithm, thereby enabling cluster-specific TIS parameter utilization. In practice, our static weight array matrix-based indexing method for parameter set lookup can be used with good results in data sets exhibiting moderate levels of 5'-complete coverage.
Conclusion: We demonstrate that improvements in statistically-based models for TIS prediction can be achieved by taking the class of each potential start-methionine into account pending certain testing conditions, and that our perceptron-based model is suitable for the TIS identification task. MetWAMer represents a well-documented, extensible, and freely available software system that can be readily re-trained for differing target applications and/or extended with existing and novel TIS prediction methods, to support further research efforts in this area.
Figures


Similar articles
-
TICO: a tool for improving predictions of prokaryotic translation initiation sites.Bioinformatics. 2005 Sep 1;21(17):3568-9. doi: 10.1093/bioinformatics/bti563. Epub 2005 Jun 30. Bioinformatics. 2005. PMID: 15994191
-
Accuracy improvement for identifying translation initiation sites in microbial genomes.Bioinformatics. 2004 Dec 12;20(18):3308-17. doi: 10.1093/bioinformatics/bth390. Epub 2004 Jul 9. Bioinformatics. 2004. PMID: 15247104
-
Computational evaluation of TIS annotation for prokaryotic genomes.BMC Bioinformatics. 2008 Mar 25;9:160. doi: 10.1186/1471-2105-9-160. BMC Bioinformatics. 2008. PMID: 18366730 Free PMC article.
-
Alternative translation start sites and hidden coding potential of eukaryotic mRNAs.Bioessays. 2008 Jul;30(7):683-91. doi: 10.1002/bies.20771. Bioessays. 2008. PMID: 18536038 Review.
-
Regulation of translation via mRNA structure in prokaryotes and eukaryotes.Gene. 2005 Nov 21;361:13-37. doi: 10.1016/j.gene.2005.06.037. Epub 2005 Oct 5. Gene. 2005. PMID: 16213112 Review.
Cited by
-
MaizeGDB becomes 'sequence-centric'.Database (Oxford). 2009;2009:bap020. doi: 10.1093/database/bap020. Epub 2009 Dec 7. Database (Oxford). 2009. PMID: 21847242 Free PMC article.
-
Dragon TIS Spotter: an Arabidopsis-derived predictor of translation initiation sites in plants.Bioinformatics. 2013 Jan 1;29(1):117-8. doi: 10.1093/bioinformatics/bts638. Epub 2012 Oct 30. Bioinformatics. 2013. PMID: 23110968 Free PMC article.
-
PreTIS: A Tool to Predict Non-canonical 5' UTR Translational Initiation Sites in Human and Mouse.PLoS Comput Biol. 2016 Oct 21;12(10):e1005170. doi: 10.1371/journal.pcbi.1005170. eCollection 2016 Oct. PLoS Comput Biol. 2016. PMID: 27768687 Free PMC article.
-
Identification of cis-regulatory sequence variations in individual genome sequences.Genome Med. 2011 Oct 10;3(10):65. doi: 10.1186/gm281. Genome Med. 2011. PMID: 21989199 Free PMC article.
-
Prediction of thermostability from amino acid attributes by combination of clustering with attribute weighting: a new vista in engineering enzymes.PLoS One. 2011;6(8):e23146. doi: 10.1371/journal.pone.0023146. Epub 2011 Aug 10. PLoS One. 2011. PMID: 21853079 Free PMC article.
References
-
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. - PubMed
-
- Preiss T, Hentze M. Starting the protein synthesis machine: eukaryotic translation initiation. BioEssays. 2003;25:1201–1211. - PubMed
-
- Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

