NFATc1 regulates PD-1 expression upon T cell activation
- PMID: 18802087
- PMCID: PMC2645436
- DOI: 10.4049/jimmunol.181.7.4832
NFATc1 regulates PD-1 expression upon T cell activation
Abstract
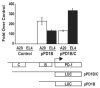
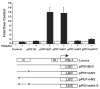
PD-1 is a transmembrane protein involved in the regulation of immunological tolerance. Multiple studies have reported an association between high levels of PD-1 expressed on T cell surfaces and exhaustion in lymphocyte populations when challenged by chronic viral infections, such as HIV. By using model systems consisting of murine EL4 cells, which constitutively express PD-1, and primary murine CD8 T cells that express PD-1 upon T cell stimulation, we have identified two tissue-specific hypersensitive sites at the 5' CR of the PD-1 locus. Gene reporter assays in CD8 T cells have shown that one of these sites has robust transcriptional activity in response to cell stimulation. Cell treatment with the calcineurin inhibitor cyclosporine A or a NFAT-specific inhibitor led to a sharp reduction in PD-1 expression in the constitutive and inducible systems. Furthermore, analysis of this region by chromatin immunoprecipitation assay revealed NFATc1 binding associated with gene activation in EL4 and primary CD8 T cells. Mutation of the NFATc1 binding site in PD-1 reporter constructs resulted in a complete loss of promoter activity. Together, these results demonstrate that PD-1 gene regulation occurs in part via the recruitment of NFATc1 to a novel regulatory element at the pdcd1 locus and provides the molecular mechanism responsible for the induction of PD-1 in response to T cell stimulation.
Conflict of interest statement
Figures
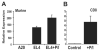





References
-
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. - PubMed
-
- Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–245. - PubMed
-
- Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006;27:195–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

