Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics
- PMID: 18804032
- PMCID: PMC2774819
- DOI: 10.1016/j.chembiol.2008.07.014
Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics
Abstract
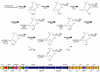
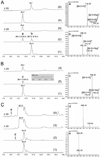
Macrolides are a class of valuable antibiotics that include a macrolactone ring, at least one appended sugar unit, and, in most cases, additional hydroxyl or epoxide groups installed by cytochrome P450 enzymes. These functional groups contribute to structural diversification and serve to improve the bioactivity profiles of natural products. Here, we have characterized in vitro two P450 enzymes from the mycinamicin biosynthetic pathway of Micromonospora griseorubida. First, MycCI was characterized as the C21 methyl hydroxylase of mycinamicin VIII, the earliest macrolide form in the postpolyketide synthase tailoring pathway. Moreover, we established that optimal activity of MycCI depends on the native ferredoxin MycCII. Second, MycG P450 catalyzes consecutive hydroxylation and epoxidation reactions with mycinamicin IV as initial substrate. These reactions require prior dimethylation of 6-deoxyallose to mycinose for effective conversion by the dual function MycG enzyme.
Figures


Similar articles
-
Function of cytochrome P450 enzymes MycCI and MycG in Micromonospora griseorubida, a producer of the macrolide antibiotic mycinamicin.Antimicrob Agents Chemother. 2012 Jul;56(7):3648-56. doi: 10.1128/AAC.06063-11. Epub 2012 Apr 30. Antimicrob Agents Chemother. 2012. PMID: 22547618 Free PMC article.
-
Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions.J Biol Chem. 2012 Nov 2;287(45):37880-90. doi: 10.1074/jbc.M112.410340. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952225 Free PMC article.
-
Production of hybrid macrolide antibiotics by exploiting the specific substrate recognition characteristics of multifunctional cytochrome P450 enzyme MycG.FEMS Microbiol Lett. 2024 Jan 9;371:fnae080. doi: 10.1093/femsle/fnae080. FEMS Microbiol Lett. 2024. PMID: 39341787
-
Genetic engineering of macrolide biosynthesis: past advances, current state, and future prospects.Appl Microbiol Biotechnol. 2010 Feb;85(5):1227-39. doi: 10.1007/s00253-009-2326-8. Epub 2009 Nov 10. Appl Microbiol Biotechnol. 2010. PMID: 19902203 Review.
-
Biosynthesis and pathway engineering of antifungal polyene macrolides in actinomycetes.J Ind Microbiol Biotechnol. 2013 Jun;40(6):529-43. doi: 10.1007/s10295-013-1258-6. Epub 2013 Mar 21. J Ind Microbiol Biotechnol. 2013. PMID: 23515854 Review.
Cited by
-
Enzymatic synthesis of indigo derivatives by tuning P450 BM3 peroxygenases.Synth Syst Biotechnol. 2023 Jul 4;8(3):452-461. doi: 10.1016/j.synbio.2023.06.006. eCollection 2023 Sep. Synth Syst Biotechnol. 2023. PMID: 37448528 Free PMC article.
-
Generation of complexity in fungal terpene biosynthesis: discovery of a multifunctional cytochrome P450 in the fumagillin pathway.J Am Chem Soc. 2014 Mar 19;136(11):4426-36. doi: 10.1021/ja500881e. Epub 2014 Mar 11. J Am Chem Soc. 2014. PMID: 24568283 Free PMC article.
-
Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.Nucleic Acids Res. 2021 Sep 20;49(16):9560-9573. doi: 10.1093/nar/gkab684. Nucleic Acids Res. 2021. PMID: 34417608 Free PMC article.
-
Reconstitution of the In Vitro Activity of the Cyclosporine-Specific P450 Hydroxylase from Sebekia benihana and Development of a Heterologous Whole-Cell Biotransformation System.Appl Environ Microbiol. 2015 Sep;81(18):6268-75. doi: 10.1128/AEM.01353-15. Epub 2015 Jul 6. Appl Environ Microbiol. 2015. PMID: 26150455 Free PMC article.
-
Structural basis of substrate specificity and regiochemistry in the MycF/TylF family of sugar O-methyltransferases.ACS Chem Biol. 2015 May 15;10(5):1340-51. doi: 10.1021/cb5009348. Epub 2015 Feb 26. ACS Chem Biol. 2015. PMID: 25692963 Free PMC article.
References
-
- Anzai Y, Saito N, Tanaka M, Kinoshita K, Koyama Y, Kato F. Organization of the biosynthetic gene cluster for the polyketide macrolide mycinamicin in Micromonospora griseorubida. FEMS Microbiol. Lett. 2003;218:135–141. - PubMed
-
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. - PubMed
-
- Chen H, Yamase H, Murakami K, Chang C-w, Zhao L, Zhao Z, Liu H-w. Expression, purification, and characterization of two N,N-dimethyltransferases, TylM1 and DesVI, involved in the biosynthesis of mycaminose and desosamine. Biochemistry. 2002;41:9165–9183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

