The role of autophagy in mammalian development: cell makeover rather than cell death
- PMID: 18804433
- PMCID: PMC2688784
- DOI: 10.1016/j.devcel.2008.08.012
The role of autophagy in mammalian development: cell makeover rather than cell death
Abstract
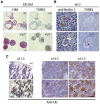
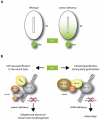
Autophagy is important for the degradation of bulk cytoplasm, long-lived proteins, and entire organelles. In lower eukaryotes, autophagy functions as a cell death mechanism or as a stress response during development. However, autophagy's significance in vertebrate development, and the role (if any) of vertebrate-specific factors in its regulation, remains unexplained. Through careful analysis of the current autophagy gene mutant mouse models, we propose that in mammals, autophagy may be involved in specific cytosolic rearrangements needed for proliferation, death, and differentiation during embryogenesis and postnatal development. Thus, autophagy is a process of cytosolic "renovation," crucial in cell fate decisions.
Figures

References
-
- Akao Y, Otsuki Y, Kataoka S, Ito Y, Tsujimoto Y. Multiple subcellular localization of bcl-2: detection in nuclear outer membrane, endoplasmic reticulum membrane, and mitochondrial membranes. Cancer Res. 1994;54:2468–2471. - PubMed
-
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved’ova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, et al. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. - PubMed
-
- Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–945. - PubMed
-
- Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J. Morphol. 1977;154:39–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

