Changing partners: moving from non-homologous to homologous centromere pairing in meiosis
- PMID: 18804891
- PMCID: PMC2861281
- DOI: 10.1016/j.tig.2008.08.006
Changing partners: moving from non-homologous to homologous centromere pairing in meiosis
Abstract
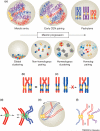
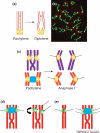
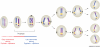
Reports of centromere pairing in early meiotic cells have appeared sporadically over the past thirty years. Recent experiments demonstrate that early centromere pairing occurs between non-homologous centromeres. As meiosis proceeds, centromeres change partners, becoming arranged in homologous pairs. Investigations of these later centromere pairs indicate that paired homologous centromeres are actively associated rather than positioned passively, side-by-side. Meiotic centromere pairing has been observed in organisms as diverse as mice, wheat and yeast, indicating that non-homologous centromere pairing in early meiosis and active homologous centromere pairing in later meiosis might be themes in meiotic chromosome behavior. Moreover, such pairing could have previously unrecognized roles in mediating chromosome organization or architecture that impact meiotic segregation fidelity.
Figures

References
-
- Bennett MD. Centromere arrangements in Triticum aestivum and their relationship to synapsis. Heredity. 1979;43:157.
-
- Brinkley BR, et al. Arrangements of kinetochores in mouse cells during meiosis and spermiogenesis. Chromosoma. 1986;94:309–317. - PubMed
-
- Church K, Moens PB. Centromere behavior during interphase and meiotic prophase in Allium fistulosum from 3-D, E.M. reconstruction. Chromosoma. 1976;56:249–263.
-
- Yunis JJ, Yasmineh WG. Model for mammalian constitutive heterocromatin. In: DuPraw FJ, editor. Advances in Cell and Molecular Biology. Academic Press; 1972. pp. 1–46.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

