Mechanism of replication-coupled DNA interstrand crosslink repair
- PMID: 18805090
- PMCID: PMC2748255
- DOI: 10.1016/j.cell.2008.08.030
Mechanism of replication-coupled DNA interstrand crosslink repair
Erratum in
- Cell.2009 May 29;137(5):972. Knipsheer, Puck [corrected to Knipscheer, Puck]
Abstract
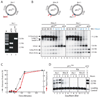
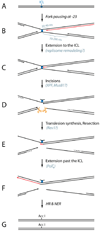
DNA interstrand crosslinks (ICLs) are toxic DNA lesions whose repair occurs in the S phase of metazoans via an unknown mechanism. Here, we describe a cell-free system based on Xenopus egg extracts that supports ICL repair. During DNA replication of a plasmid containing a site-specific ICL, two replication forks converge on the crosslink. Subsequent lesion bypass involves advance of a nascent leading strand to within one nucleotide of the ICL, followed by incisions, translesion DNA synthesis, and extension of the nascent strand beyond the lesion. Immunodepletion experiments suggest that extension requires DNA polymerase zeta. Ultimately, a significant portion of the input DNA is fully repaired, but not if DNA replication is blocked. Our experiments establish a mechanism for ICL repair that reveals how this process is coupled to DNA replication.
Figures





References
-
- Akkari YM, Bateman RL, Reifsteck CA, D'Andrea AD, Olson SB, Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol Genet Metab. 2001;74:403–412. - PubMed
-
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. - PubMed
-
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278:5250–5254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

