Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends
- PMID: 18805091
- PMCID: PMC2662516
- DOI: 10.1016/j.cell.2008.08.037
Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends
Abstract
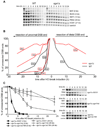
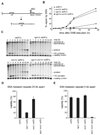
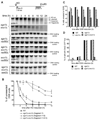
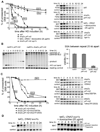
Formation of single-strand DNA (ssDNA) tails at a double-strand break (DSB) is a key step in homologous recombination and DNA-damage signaling. The enzyme(s) producing ssDNA at DSBs in eukaryotes remain unknown. We monitored 5'-strand resection at inducible DSB ends in yeast and identified proteins required for two stages of resection: initiation and long-range 5'-strand resection. We show that the Mre11-Rad50-Xrs2 complex (MRX) initiates 5' degradation, whereas Sgs1 and Dna2 degrade 5' strands exposing long 3' strands. Deletion of SGS1 or DNA2 reduces resection and DSB repair by single-strand annealing between distant repeats while the remaining long-range resection activity depends on the exonuclease Exo1. In exo1Deltasgs1Delta double mutants, the MRX complex together with Sae2 nuclease generate, in a stepwise manner, only few hundred nucleotides of ssDNA at the break, resulting in inefficient gene conversion and G2/M damage checkpoint arrest. These results provide important insights into the early steps of DSB repair in eukaryotes.
Figures


References
-
- Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. - PubMed
-
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. - PubMed
-
- Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. - PubMed
-
- Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

