Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation
- PMID: 18805092
- PMCID: PMC2628631
- DOI: 10.1016/j.cell.2008.07.022
Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation
Abstract
Cullin-RING ligases (CRLs) comprise the largest ubiquitin E3 subclass, in which a central cullin subunit links a substrate-binding adaptor with an E2-binding RING. Covalent attachment of the ubiquitin-like protein NEDD8 to a conserved C-terminal domain (ctd) lysine stimulates CRL ubiquitination activity and prevents binding of the inhibitor CAND1. Here we report striking conformational rearrangements in the crystal structure of NEDD8~Cul5(ctd)-Rbx1 and SAXS analysis of NEDD8~Cul1(ctd)-Rbx1 relative to their unmodified counterparts. In NEDD8ylated CRL structures, the cullin WHB and Rbx1 RING subdomains are dramatically reoriented, eliminating a CAND1-binding site and imparting multiple potential catalytic geometries to an associated E2. Biochemical analyses indicate that the structural malleability is important for both CRL NEDD8ylation and subsequent ubiquitination activities. Thus, our results point to a conformational control of CRL activity, with ligation of NEDD8 shifting equilibria to disfavor inactive CAND1-bound closed architectures, and favor dynamic, open forms that promote polyubiquitination.
Figures
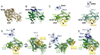
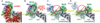



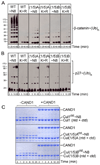
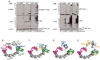
Comment in
-
A ubiquitin-like protein unleashes ubiquitin ligases.Cell. 2008 Oct 17;135(2):209-11. doi: 10.1016/j.cell.2008.09.049. Cell. 2008. PMID: 18957195
References
-
- Amir RE, Iwai K, Ciechanover A. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J Biol Chem. 2002;277:23253–23259. - PubMed
-
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. - PubMed
-
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Bio. 1999;1:438–443. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

