Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis
- PMID: 18805909
- PMCID: PMC2576632
- DOI: 10.1093/jxb/ern234
Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis
Abstract
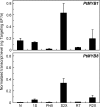
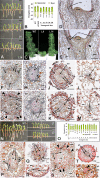
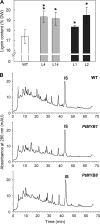
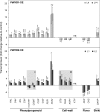
The involvement of two R2R3-MYB genes from Pinus taeda L., PtMYB1 and PtMYB8, in phenylpropanoid metabolism and secondary cell wall biogenesis was investigated in planta. These pine MYBs were constitutively overexpressed (OE) in Picea glauca (Moench) Voss, used as a heterologous conifer expression system. Morphological, histological, chemical (lignin and soluble phenols), and transcriptional analyses, i.e. microarray and reverse transcription quantitative PCR (RT-qPCR) were used for extensive phenotyping of MYB-overexpressing spruce plantlets. Upon germination of somatic embryos, root growth was reduced in both transgenics. Enhanced lignin deposition was also a common feature but ectopic secondary cell wall deposition was more strongly associated with PtMYB8-OE. Microarray and RT-qPCR data showed that overexpression of each MYB led to an overlapping up-regulation of many genes encoding phenylpropanoid enzymes involved in lignin monomer synthesis, while misregulation of several cell wall-related genes and other MYB transcription factors was specifically associated with PtMYB8-OE. Together, the results suggest that MYB1 and MYB8 may be part of a conserved transcriptional network involved in secondary cell wall deposition in conifers.
Figures
References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300.
-
- Björkmann A. Studies on finely divided wood. 1. Extraction of lignin with neutral solvents. Suen Papperstidn. 1956;59:477.
-
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Reviews in Plant Biology. 2003;54:519–546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

