MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile
- PMID: 18805954
- PMCID: PMC2577246
- DOI: 10.1104/pp.108.121913
MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile
Abstract
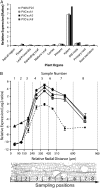
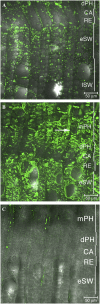
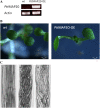
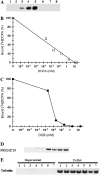
We have identified a gene, denoted PttMAP20, which is strongly up-regulated during secondary cell wall synthesis and tightly coregulated with the secondary wall-associated CESA genes in hybrid aspen (Populus tremula x tremuloides). Immunolocalization studies with affinity-purified antibodies specific for PttMAP20 revealed that the protein is found in all cell types in developing xylem and that it is most abundant in cells forming secondary cell walls. This PttMAP20 protein sequence contains a highly conserved TPX2 domain first identified in a microtubule-associated protein (MAP) in Xenopus laevis. Overexpression of PttMAP20 in Arabidopsis (Arabidopsis thaliana) leads to helical twisting of epidermal cells, frequently associated with MAPs. In addition, a PttMAP20-yellow fluorescent protein fusion protein expressed in tobacco (Nicotiana tabacum) leaves localizes to microtubules in leaf epidermal pavement cells. Recombinant PttMAP20 expressed in Escherichia coli also binds specifically to in vitro-assembled, taxol-stabilized bovine microtubules. Finally, the herbicide 2,6-dichlorobenzonitrile, which inhibits cellulose synthesis in plants, was found to bind specifically to PttMAP20. Together with the known function of cortical microtubules in orienting cellulose microfibrils, these observations suggest that PttMAP20 has a role in cellulose biosynthesis.
Figures




Similar articles
-
Evolution of a domain conserved in microtubule-associated proteins of eukaryotes.Adv Appl Bioinform Chem. 2008;1:51-69. doi: 10.2147/aabc.s3211. Epub 2008 Sep 23. Adv Appl Bioinform Chem. 2008. PMID: 21918606 Free PMC article.
-
Differential expression patterns of two cellulose synthase genes are associated with primary and secondary cell wall development in aspen trees.Planta. 2004 Nov;220(1):47-55. doi: 10.1007/s00425-004-1329-z. Epub 2004 Jul 20. Planta. 2004. PMID: 15278458
-
Cloning and characterization of cellulose synthase-like gene, PtrCSLD2 from developing xylem of aspen trees.Physiol Plant. 2004 Apr;120(4):631-641. doi: 10.1111/j.0031-9317.2004.0271.x. Physiol Plant. 2004. PMID: 15032825
-
Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins.Curr Opin Plant Biol. 2013 Dec;16(6):743-8. doi: 10.1016/j.pbi.2013.10.010. Epub 2013 Nov 7. Curr Opin Plant Biol. 2013. PMID: 24210792 Review.
-
Secondary cell wall patterning during xylem differentiation.Curr Opin Plant Biol. 2012 Feb;15(1):38-44. doi: 10.1016/j.pbi.2011.10.005. Epub 2011 Nov 9. Curr Opin Plant Biol. 2012. PMID: 22078063 Review.
Cited by
-
Arabidopsis XTH4 and XTH9 Contribute to Wood Cell Expansion and Secondary Wall Formation.Plant Physiol. 2020 Apr;182(4):1946-1965. doi: 10.1104/pp.19.01529. Epub 2020 Jan 31. Plant Physiol. 2020. PMID: 32005783 Free PMC article.
-
The Cytoskeleton and Its Role in Determining Cellulose Microfibril Angle in Secondary Cell Walls of Woody Tree Species.Plants (Basel). 2020 Jan 10;9(1):90. doi: 10.3390/plants9010090. Plants (Basel). 2020. PMID: 31936868 Free PMC article. Review.
-
Comparative transcriptome analyses of fruit development among pears, peaches, and strawberries provide new insights into single sigmoid patterns.BMC Plant Biol. 2020 Mar 6;20(1):108. doi: 10.1186/s12870-020-2317-6. BMC Plant Biol. 2020. PMID: 32143560 Free PMC article.
-
Cellulose biosynthesis inhibitors: comparative effect on bean cell cultures.Int J Mol Sci. 2012;13(3):3685-3702. doi: 10.3390/ijms13033685. Epub 2012 Mar 20. Int J Mol Sci. 2012. PMID: 22489176 Free PMC article.
-
A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells.Nat Cell Biol. 2011 Jul 17;13(8):973-80. doi: 10.1038/ncb2294. Nat Cell Biol. 2011. PMID: 21765420
References
-
- Abe H, Funada R, Imaizumi H, Ohtani J, Fukazawa K (1995) Dynamic changes in the arrangement of cortical microtubules in conifer tracheids during differentiation. Planta 197 418–421
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 - PubMed
-
- An G (1987) Binary ti-vectors for plant transformation and promoter analysis. Methods Enzymol 153 292–305
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

