Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code
- PMID: 18805961
- PMCID: PMC2682356
- DOI: 10.1182/blood-2008-05-157560
Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code
Abstract
Endoglin is an accessory receptor for TGF-beta signaling and is required for normal hemangioblast, early hematopoietic, and vascular development. We have previously shown that an upstream enhancer, Eng -8, together with the promoter region, mediates robust endothelial expression yet is inactive in blood. To identify hematopoietic regulatory elements, we used array-based methods to determine chromatin accessibility across the entire locus. Subsequent transgenic analysis of candidate elements showed that an endothelial enhancer at Eng +9 when combined with an element at Eng +7 functions as a strong hemato-endothelial enhancer. Chromatin immunoprecipitation (ChIP)-chip analysis demonstrated specific binding of Ets factors to the promoter as well as to the -8, +7+9 enhancers in both blood and endothelial cells. By contrast Pu.1, an Ets factor specific to the blood lineage, and Gata2 binding was only detected in blood. Gata2 was bound only at +7 and GATA motifs were required for hematopoietic activity. This modular assembly of regulators gives blood and endothelial cells the regulatory freedom to independently fine-tune gene expression and emphasizes the role of regulatory divergence in driving functional divergence.
Figures
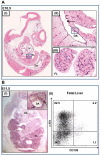
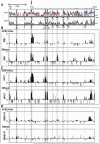
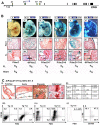
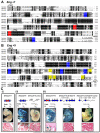
References
-
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. - PubMed
-
- Donaldson IJ, Chapman M, Kinston S, et al. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum Mol Genet. 2005;14:595–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

