Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications
- PMID: 18805973
- PMCID: PMC2583624
- DOI: 10.1128/JB.00984-08
Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications
Abstract
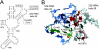
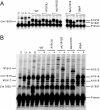
Translocation during the elongation phase of protein synthesis involves the relative movement of the 30S and 50S ribosomal subunits. This movement is the target of tuberactinomycin antibiotics. Here, we describe the isolation and characterization of mutants of Thermus thermophilus selected for resistance to the tuberactinomycin antibiotic capreomycin. Two base substitutions, A1913U and mU1915G, and a single base deletion, DeltamU1915, were identified in helix 69 of 23S rRNA, a structural element that forms part of an interribosomal subunit bridge with the decoding center of 16S rRNA, the site of previously reported capreomycin resistance base substitutions. Capreomycin resistance in other bacteria has been shown to result from inactivation of the TlyA methyltransferase which 2'-O methylates C1920 of 23S rRNA. Inactivation of the tlyA gene in T. thermophilus does not affect its sensitivity to capreomycin. Finally, none of the mutations in helix 69 interferes with methylation at C1920 or with pseudouridylation at positions 1911 and 1917. We conclude that the resistance phenotype is a consequence of structural changes introduced by the mutations.
Figures


References
-
- Ali, I. K., L. Lancaster, J. Feinberg, S. Joseph, and H. F. Noller. 2006. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell 23865-874. - PubMed
-
- Cameron, D. M., J. Thompson, P. E. March, and A. E. Dahlberg. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 31927-35. - PubMed
-
- Dallas, A., and H. F. Noller. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8855-864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

