Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions
- PMID: 18805977
- PMCID: PMC2576672
- DOI: 10.1128/JB.01002-08
Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions
Abstract
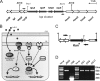
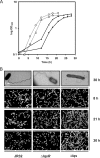
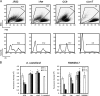
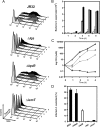
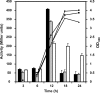
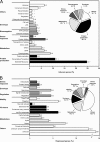
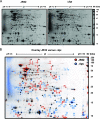
The causative agent of Legionnaires' disease, Legionella pneumophila, is a natural parasite of environmental protozoa and employs a biphasic life style to switch between a replicative and a transmissive (virulent) phase. L. pneumophila harbors the lqs (Legionella quorum sensing) cluster, which includes genes encoding the autoinducer synthase LqsA, the sensor kinase LqsS, the response regulator LqsR, and a homologue of HdeD, which is involved in acid resistance in Escherichia coli. LqsR promotes host-cell interactions as an element of the stationary-phase virulence regulatory network. Here, we characterize L. pneumophila mutant strains lacking all four genes of the lqs cluster or only the hdeD gene. While an hdeD mutant strain did not have overt physiological or virulence phenotypes, an lqs mutant showed an aberrant morphology in stationary growth phase and was defective for intracellular growth, efficient phagocytosis, and cytotoxicity against host cells. Cytotoxicity was restored upon reintroduction of the lqs genes into the chromosome of an lqs mutant strain. The deletion of the lqs cluster caused more-severe phenotypes than deletion of only lqsR, suggesting a synergistic effect of the other lqs genes. A transcriptome analysis indicated that in the stationary phase more than 380 genes were differentially regulated in the lqs mutant and wild-type L. pneumophila. Genes involved in protein production, metabolism, and bioenergetics were upregulated in the lqs mutant, whereas genes encoding virulence factors, such as effectors secreted by the Icm/Dot type IV secretion system, were downregulated. A proteome analysis revealed that a set of Icm/Dot substrates is not produced in the absence of the lqs gene cluster, which confirms the findings from DNA microarray assays and mirrors the virulence phenotype of the lqs mutant strain.
Figures

References
-
- Albers, U., K. Reus, H. A. Shuman, and H. Hilbi. 2005. The amoebae plate test implicates a paralogue of lpxB in the interaction of Legionella pneumophila with Acanthamoeba castellanii. Microbiology 151167-182. - PubMed
-
- Albers, U., A. Tiaden, T. Spirig, D. Al Alam, S. M. Goyert, S. C. Gangloff, and H. Hilbi. 2007. Expression of Legionella pneumophila paralogous lipid A biosynthesis genes under different growth conditions. Microbiology 1533817-3829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

